One of the central archetypes of clinical practice is the mystery of a patient who is clearly sick yet lacks a definitive diagnosis. With infectious disease, the near-infinite diversity of pathogens makes this dilemma a unique challenge. The advent of tests that perform rapid genetic sequencing to identify nonhuman, microbial genomic information (also known as metagenomic next-generation sequencing, or mNGS) offers a singular appeal in this setting: The genetic identity of the pathogen could provide an immediate answer to the central mystery of the infected patient. Unfortunately, the cost of mNGS is typically 10–20 times that of most conventional laboratory tests for infectious disease.
How can clinicians and labs deploy mNGS to maximize diagnostic yield while minimizing cost? We offer three perspectives from key stakeholders, as well as a theoretical framework for optimizing clinical benefit and laboratory stewardship.
The Microbiologist’s Perspective
The technology underlying mNGS is still in its early stages. Conventional microbiology testing typically provides an array of information beyond a singular positive or negative signal: The isolate is either viable or not viable, the relative abundance of an organism can be compared with normal flora or signals of contamination, and phenotypic susceptibility testing provides basis for treatment. By contrast, mNGS offers essentially binary diagnostic information about all organisms, independent of either mechanism or clinical hypothesis. The implications of each result may not be fully known, and often there are no conventional tests to serve as adequate comparators.
In the United States, mNGS testing is almost entirely limited to two reference labs—Karius, Inc. and the University of California, San Francisco (UCSF). Karius provides a turnaround time of just a few days, but the analysis is limited to DNA. UCSF includes both RNA and DNA, although the time to result is advertised as 1−2 weeks. Perhaps the most significant limitation of each is that the specimen types are restricted to plasma and cerebrospinal fluid (CSF), respectively.
A key tenet of microbiological diagnosis is that the highest yield is obtained when sampling at the site of primary infection and at the time of maximal organism burden. In practice, labs frequently deploy mNGS when these conditions are not met, for instance when a direct specimen is difficult to obtain or long after antimicrobial treatment has been initiated. Study of plasma mNGS has spanned myriad clinical indications from pneumonia to soft tissue infection, and there is no consensus as to which criteria should serve as triggers to order the test. On the other hand, mNGS of CSF has been applied to a limited number of disease states and accompanied by familiar supplementary markers of central nervous system infection—making it amenable to preliminary, but straightforward, laboratory policy.
The Clinician’s Perspective
Available data indicate that mNGS can augment conventional testing for the diagnosis of infectious pathogens but should not replace it. In a study including 41 patients with encephalitis due to an established infectious disease, the UCSF mNGS assay identified the same pathogen as conventional testing in 32% of patients and identified a pathogen that was not detected with conventional testing in 22% of patients; however, mNGS missed a pathogen that was found on conventional testing in 45% of patients (N Engl J Med 2019; doi: 10.1056/NEJMoa1803396).This demonstrates a fundamental problem for clinicians: Without a gold standard of diagnostic truth, it is difficult to calculate the clinical sensitivity or specificity of mNGS testing.
Furthermore, the exquisite sensitivity of NGS may lead to false-positive identification of commensal organisms and unlikely pathogens. In an initial evaluation of the Karius test in 348 patients presenting with symptoms of sepsis, 18% had a positive result by mNGS alone (not detected by conventional testing) in which the pathogen was felt to be an “unlikely” (9%) or “possible” (9%) cause of infection (Nat Microbiol 2019; doi: 10.1038/s41564-018-0349-6). This places clinicians in the difficult position of determining what represents a true infection.
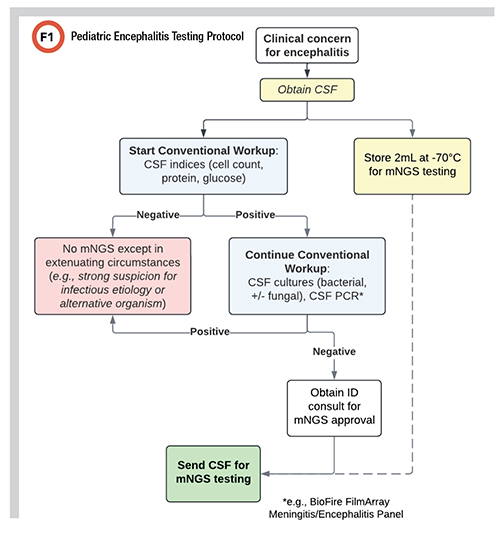
Faced with uncertain clinical utility and high cost, providers in our institution frequently deploy mNGS as a last resort for patients whose condition is deteriorating despite broad-spectrum antimicrobial treatment. However, clinical evidence demonstrates that mNGS has the highest likelihood of detecting a pathogen during the early phase of illness (J Infect 2019; doi: 10.1016/j.jinf.2018.12.001). To address this, we developed a protocol for pediatric encephalitis that encourages collecting a sample for mNGS during the acute phase of infection, followed by frozen storage while awaiting the results of initial testing via conventional methods (Figure 1).
The Laboratory Stewardship Perspective
A significant barrier to mNGS implementation in hospitals is expense: One test can cost $2,000−$2,500 (Karius and UCSF, respectively)*. However, the diagnostic evaluation for encephalitis frequently leads to significant laboratory, medication, and hospital-based costs: Any single test obtained during our diagnostic evaluation typically costs $65−$560 (median: $181).
We conducted an analysis comparing costs of diagnostic evaluation with and without mNGS. Our infectious disease division pursues a tiered approach to the diagnosis of encephalitis, in which the bundled costs of each tier of evaluation typically amount to approximately $3,000 for laboratory studies. This is similar to the cost for mNGS, and for additional context, would be similar in scale to the costs of an MRI of the head at our institution. Altogether, testing costs, with or without mNGS, are far less than the estimated cost of a day of hospitalization in the intensive care unit ($16,000) or the estimated cost of a brain biopsy ($46,500)—the test of last resort in encephalitis workup.
The protocol we propose accounts for this rough cost equivalency, which suggests that mNGS is worth sending if it could avoid significant additional testing or brain biopsy, or shorten hospitalization by even 1 day. If an mNGS result obviates the need for a brain biopsy just one in every 25 cases, cost equivalency is reached—and this is without considering the incalculable value of avoiding such a procedure.
The Future of mNGS in Infectious Disease
To optimally implement mNGS, it is necessary to know how often the results lead to cost savings like these. For instance, one cost-benefit model for fever of unknown origin estimated that mNGS of plasma would have to provide actionable results 60% of the time to break even (PLoS One 2019; doi:10.1371/journal.pone.0194648). Currently, there are few economic evaluations of mNGS for infectious diseases in the literature, and additional research is needed.
More importantly, clinicians require tools that help identify which patients are likely to benefit from mNGS. One approach would be a clinical scoring tool similar to the Neonatal Sepsis Calculator for evaluating the likelihood of serious bacterial infection based on a set of validated clinical markers (JAMA Pediatr 2017; doi: 10.1001/jamapediatrics.2016.4678). We believe that future research should focus on this type of analysis, testing clinical prediction rules to determine the expected benefit from mNGS.
By establishing quantitative measures of clinical utility and combining these with transparent data about costs, laboratory stewardship can empower providers to deploy testing in the most efficient pursuit of diagnostic certainty.
* Costs are based on the self-pay rate from published Standard Charges data for Seattle Children’s Hospital for single tests. Calculations of estimated per diem costs of hospitalization and procedure costs were made by the Seattle Children’s Hospital department of finance and business management.
Caleb Stokes, MD, PhD, is acting assistant professor, infectious disease, at the University of Washington and Seattle Children’s Hospital in Seattle. +Email: [email protected]
Erin Chung, MD, is a pediatric infectious disease fellow at the University of Washington and Seattle Children’s Hospital in Seattle. +Email: [email protected]
Drew Bell, PhD, is codirector of the microbiology lab at Seattle Children’s Hospital and clinical assistant professor at the University of Washington in Seattle. +Email: [email protected]