Advances in machine learning and data analytics are transforming the field of laboratory medicine. AACC has focused on enabling members to lead at the forefront of this evolution in healthcare, using the wealth of laboratories’ data to create better medical outcomes for patients.
As a pillar in the association’s strategic plan and through the actions of the Data Analytics Steering Committee, AACC has formed a three-pronged approach for data analytics education, data access, and community building. As part of this effort, AACC worked with the informatics section in the department of pathology and immunology of Washington University School of Medicine, St. Louis (WUSM), to introduce AACC’s first machine-learning challenge, the Predicting PTHrP Results Competition, at the 2022 AACC Annual Scientific Meeting in Chicago.
Through this competition,AACC and WUSM aimed to engage the community of laboratory medicine practitioners in a fun and friendly online environment where they could practice their data analytics skills, learn from each other, and see how others approach problems on the data-driven side of laboratory medicine.
“With data analytics, we’re trying to bring more quantitative rigor to lab medicine—as you’d see in other disciplines, like finance,” said competition organizer Mark Zaydman, MD, PhD, an assistant professor of pathology and immunology at WUSM. “Ultimately, we want to achieve better outcomes for the patient and the institution. That’s the outlook we brought to the call for submissions to the PTHrP results competition.”
Leveraging Real-World Data
The purpose of the competition was to see if a machine-learning approach could better predict test outcomes compared to the traditional manual approach many clinical laboratories use, reviewing calcium and PTH results to identify potential inappropriate parathyroid hormone-related peptide (PTHrP) orders.
Clinicians rely on PTHrP measurement to help establish a diagnosis of humoral hypercalcemia of malignancy—a rare form of cancer that causes, among other things, high levels of calcium in the blood. The problem: Clinicians often order it for patients with low pretest probability. Excessive PTHrP testing can lead to expensive, unnecessary, and potentially harmful procedures, including invasive laboratory testing to locate a possibly nonexistent cancerous tumor. A successful predictive algorithm would help laboratorians quickly and accurately identify potentially inappropriate PTHrP test orders by predicting whether laboratory data available at the time of order already suggest an abnormal PTHrP result.
Competition participants formed teams and used securely shared real, deidentified clinical data from PTHrP orders at WUSM to build their predictive algorithms. This is termed the “practice dataset”. Using real clinical data was a big deal because most machine-learning competitions use synthesized datasets, Zaydman noted. “We wanted the competition to be grounded in something that was realistic because it’s a better way to learn,” he said. “With real patient data, the process is going to be more engaging and more interesting.”
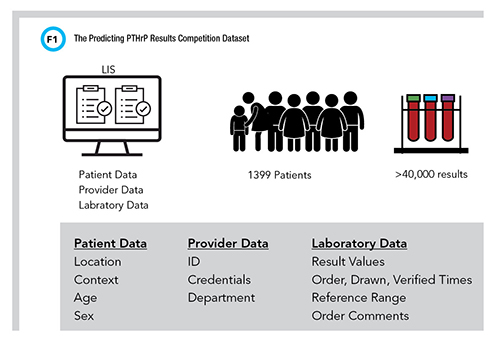
Organizers set up the competition using Kaggle, a popular online platform for machine-learning modeling and contests, and selected F1 score (the harmonic mean of sensitivity and specificity) as the performance metric (Figure 1).
A major challenge for the teams was developing a predictive model that achieved high accuracy without overfitting it to the public dataset (the practice dataset). Overfitting would mean the algorithm worked well on the initial data but failed if applied to new data and was not generalizable. Organizers used a second, private dataset to judge the algorithm’s effectiveness.
From May to June 2022, 24 teams ran a total of 395 iterations of their predictive models through the public dataset. Each time a team submitted a predictive model for an attempt, they used the resulting F1 score to improve—or “train”—the model. For the final attempt, each team ran their predictive model through the private dataset.
How the “Dream Team” Won
The winning team, Team Kagglist, achieved an F1 score of 0.9 with their predictive model. For reference, WUSM’s manual approach for identifying patients at risk for PTHrP had an F1 score of 0.6, making the algorithm a significant improvement over standard practice.
Team Kagglist was an interdisciplinary group of experts in computer science, laboratory medicine, and healthcare epidemiology, earning them the moniker “dream team” from Zaydman.
One of the team’s strong points was its strategy for preventing overfitting their model while maximizing predictive performance. According to team lead Fei Wang, PhD, they approached the competition as a classification problem and then tested different kinds of models. “There was no magic involved in preventing overfitting,” said Wang. “We avoided over-complicated models, like deep learning, which would be overparameterized for this problem, and used standard machine-learning practice with an emphasis on adaptation to refine our algorithm.”
In his work as associate professor of health informatics at Weill Cornell Medicine, New York, Wang said he often emphasizes adaptation with predictive models like the one he and his teammates created for this competition. “We shouldn’t expect a predictive model trained on data from one hospital to automatically work at other hospitals,” he said. “Ultimately, we should aim to create adaptive models that can be fine-tuned by other institutions for their specific populations.”
Wang said Kaggle competitions are a great way for clinical laboratorians to learn more about data analytics. “The whole deep learning phenomenon came from a competition organized by the computer vision community,” he noted. “The competition format is a very good way to get engagement from people from different places and to initiate new collaborations.”
The competition concluded with a presentation and discussion led by Team Kaggle’s Yingheng Wang at the AACC Informatics Division Annual Meeting during the AACC Annual Scientific Meeting & Clinical Lab Expo on July 25, 2022.
The Data Analytics Steering Committee is looking forward to more such competitions at future AACC Annual Scientific Meetings. “The quality of all 24 models was excellent and showed a high degree of accuracy for the very difficult task we challenged participants with,” Zaydman said. “This competition really showed our community is ready to engage with sophisticated machine learning and data analytics tools.”
A Window on the Future of Laboratory Medicine
Regardless of their role in the laboratory, data analytics skills allow laboratorians to speak the same language as their informatics and computer science colleagues. Moreover, existing tools and software already are demonstrating that they’re powerful enough to make a difference in improving patient care.
Computer modeling requires collaboration between domain experts. Teams need people who can not only explain the assays and clinical problem but also understand data structure and identify potential pitfalls when working with clinical data.
To help members develop expertise in data analytics, AACC and the Data Analytics Steering Committee plan to expand training resources and are compiling educational resources on opportunities in data analytics in laboratory medicine.
“AACC has put forward that data analytics is a key area for growth and development within the medical laboratorian profession,” said chair of the AACC Data Analytics Steering Committee Patrick Mathias, MD, PhD. “Laboratories generate large amounts of data—with advances in machine learning, we have the opportunity to learn from that data and improve patient outcomes.”
Zaydman sees current advances in data analytics as just the beginning. “The future of medicine is going to be an integration of human intelligence and computer intelligence,” Zaydman predicted. “We’re at the very outset of this evolution—it’s an exciting time to be in laboratory medicine!”
Sarah Michaud is a freelance writer who lives in London. +Email: [email protected]
The Champions: