Nuclear magnetic resonance (NMR) spectroscopy has a storied history. Edward Purcell and Felix Bloch developed the NMR technology in the late 1940s, for which they earned a Nobel prize in physics. The principle underlying this approach is that radio waves excite intramolecular magnetic fields around atomic nuclei, resulting in chemical shifts and changes in resonance frequency. These changes in resonance are akin to how potassium or sodium give off different wavelengths of light when heated in flame emission photometry.
The earliest applications of NMR were based on the chemical shift that occurs in the process and the ability to use the resulting data to elucidate the structures of complex molecules. Imaging applications followed that exploited the spatial resolution of molecules using magnetic resonance imaging (MRI) platforms. A third area of NMR takes advantage of nuclear spin relaxation rates. In this last category, called NMR relaxometry, the available testing devices have evolved into benchtop devices and even portable configured ones.
Current Uses of NMR
Clinical laboratories make quite limited use of NMR in comparison with methodologies like mass spectrometry and UV-visible light spectroscopy; however, medical imaging departments of major medical centers have adopted NMR widely.
Early versions of MRI were largely designed to provide tissue or organ imaging due to sensitivity limitations associated with other technologies, including spectral analysis. However, later versions of the platform with stronger magnetics allowed MRI to also produce magnetic resonance spectroscopy (MRS) information and provide qualitative and semiquantitative resolution for the various molecules present. These achievements expanded our capacity to evaluate metabolic and physiologic aspects of the body.
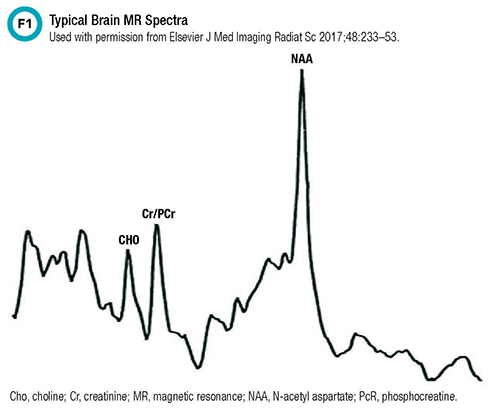
One area in which MRS has shown particular promise is in the diagnosis of congenital metabolic disorders. Given the large number of potential metabolites in the body and the high inter-individual variability in analyte concentrations among patient populations, MRS offers useful data. For example, within the brain, MRS recognizes metabolites such as N-acetyl aspartate, creatine, choline, myoinositol, lactate, glutamate, and glutamine molecules. Figure 1 illustrates the typical spectra seen in a healthy brain.
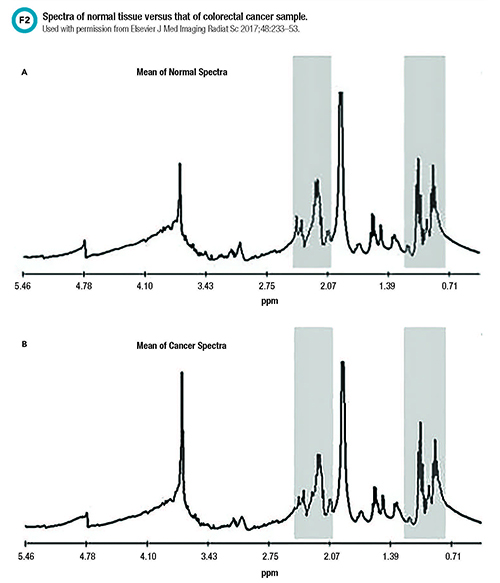
In addition to its use in discerning congenital metabolic disorders, MRS is very useful in characterizing the metabolism of tumor cells and inflammatory conditions. MRS has been used to detect the presence of brain, breast, and gastrointestinal tumors (Figure 2). Neuropsychiatric applications of MRS in evaluating systemic lupus erythematosus and multiple sclerosis, in targeting brain areas for treating epilepsy, and in predicting risk for Alzheimer’s disease and dementia also have been described in the literature.
While these applications are still adjuncts to more traditional diagnostic tools, our colleagues in radiology and medical imaging have taken full advantage of NMR to offer combined imaging and physiologic/pathophysiology assessments using a noninvasive approach.
NMR in Clinical Laboratories
Despite its potential being noted for more than 4 decades in the literature, NMR has not been broadly implemented within clinical laboratories. Today this promising technology is used mainly to quantify and characterize lipoproteins and lipids, along with some qualitative applications in microbiology.
Throughout the literature authors have noted that analytical NMR should be cost competitive with existing automated analyzers, considering there are no reagents and that a single NMR spectrum could replace a dozen or so chemical analyses. But the clinical utilization reviews for this technology still primarily discuss as its main application: imaging of soft tissues—liver, heart, brain, and kidneys.
While not as commonplace as comprehensive metabolic panel testing, NMR technology helps determine specialized parameters for lipoprotein and lipid testing. Publications describing its use in clinical laboratories began to appear in the literature in the 1980s. NMR spectroscopy can be used to measure the size and density of lipoprotein molecules, and the data it generates improves the risk assessment for atherosclerotic cardiovascular disease compared with low-density lipoprotein cholesterol levels. However, even after a few decades of research and development, use of NMR for lipoprotein profiling still lacks standardization and comparability to other methods. A 2018 article published in Clinical Chemistry noted that NMR-based measurements lacked comparability, even within the technology, and that the software used for data analysis was likely part of the problem.
In addition to its role in specialized lipoprotein measurements, NMR has been explored for point-of-care testing. Researchers have proposed using magnetic nanoparticles to enhance or amplify the signal, called microNMR, in order to bring NMR to bedsides or clinics. In fact, in 2014 the Food and Drug Administration approved an NMR-based platform called the T2Dx, from T2Biosystems, with one panel for identifying sepsis-causing bacterial infections and another for fungal pathogens in whole blood. These platforms are capable of detecting the organisms within 3-5 hours of test initiation. Researchers are also working on applications for detecting Lyme disease and carbapenem-resistant organisms.
The same T2Dx NMR benchtop device has been used to demonstrate proof-of-concept testing for several hemostatic parameters. Changes based on the relaxation of molecules offer information on clotting, clot retraction, and fibrinolytic processes. Additional applications that provide hematocrit, platelet activity, and fibrinogen levels have been described. Similar to applications in microbiology, benchtop-based NMR assays provide comparable data to more traditional coagulation assays, but with significantly less sample (<50uL) and with start-to-result times on the order of 15 minutes versus 30-150 minutes for traditional assays.
Historically, analysis of inborn errors of metabolism has been based on a targeted approach to diagnostic workflows. Research, though, typically utilizes more untargeted processes in order to take advantage of the deep datasets provided by NMR-based analysis. The evolution or translation of these research findings into clinically actionable data has yet to occur. Tebani and colleagues have used the phrase “the curse of dimensionality” to describe the issue of transforming NMR-based studies’ complex, highly dimensional datasets into the more targeted questions typical in the differential diagnosis of a metabolic disease.
Decades of incomplete and poorly designed epidemiologic and biomarker discovery studies have created misconceptions about the power of NMR in metabolomics. As such, the dialogue surrounding the clinical application of NMR spectroscopy for metabolic disease testing still commonly involves the terms “may be useful” or “underappreciated.” Yet, the commercially available applications in lipid profiling and microbiology demonstrate the technology not only has potential but also clinical applicability.
To date, other than assessing lipid metabolism, applications of NMR in metabolic disease testing remain in the whole body/tissue imaging arena. NMR-based testing of blood and other fluids is largely stuck in discovery, translational, or proof-of-concept phases. Several authors have put forth rationales to explain NMR’s lack of uptake, and the common thread appears to be data analysis and informatics limitations. Combined with a paradigm shift moving from targeted to untargeted screening workflows and the issues with harmonization of results, it’s not unreasonable to see why NMR still has not become mainstream.
Manuscripts published in Clinical Chemistry in the 1960s and 70s described applications of NMR in chemistry laboratories, and a 1984 review included the comment that “routine in vitro use of analytical NMR spectrometers is likely to be very important to the pathologist’s role in diagnosis with NMR imaging/spectroscopy.” But jump forward 30 years and an editorial in Clinical Chemistry noted that “modern NMR platforms are a powerful tool to reveal potential new biomarkers for disease risk assessment and clinical applications.” The editorial goes on to discuss the limitations of NMR spectral data analysis techniques that constrain their ability to provide absolute quantification. But the author also indicated that NMR utilization was hampered by the lack of substantial amounts of epidemiological and clinical studies and the data to support the clinical utility of NMR-based metabolic quantification(s).
The editorialist cited an associated manuscript by Otvos and colleagues in that same issue of Clinical Chemistry as an example of a different NMR data analysis approach that demonstrated the technology’s ability to provide absolute quantification in line with usual clinical laboratory expectations. Fast forward 3 years to 2018 and Ala-Korpela, in a Clinical Chemistry perspective, still lacked confidence in getting NMR- based metabolomics to the clinical space, citing as factors no clear criteria for analysis of NMR data and no standards for reporting the data. The author concluded that one of the key issues about disseminating NMR in clinical lab practice was to minimize the hype.
Conclusion
Clearly the potential of NMR-based technology has only improved over the decades but still appears underutilized and under researched. As the metabolome, metabolomics, and personalized medicine become hotter topics in clinical diagnostics, clinical laboratorians need to keep the capabilities of NMR in mind when looking for technology to address the questions being asked and improvements in efficiencies being demanded.
Mark Kellogg PhD, DABCC, FADLM, is the associate director of chemistry at Boston Children’s Hospital and assitant professor of pathology at Harvard Medical School in Boston. +Email: mark.kellogg@childrens.harvard.edu
Suggested Readings
- Ala-Korpela M. Serum nuclear magnetic resonance spectroscopy: one more step toward clinical utility. Clin Chem 2015; 61: 681-3.
- Ala-Korpela M. Objective metabolomics research. Clin Chem 2018;64:30-3.
- Ala-Korpela M, Davey Smith G. Metabolic profiling-multitude of technologies with great research potential, but (when) will translation emerge? Int J Epidemiol 2016;45:1311-8.
- Bock JL. NMR in clinical chemistry—where do we stand? Clin Chem 1994;40:1215-7.
- Bottomley PA, Hardy CJ. Rapid, reliable in vivo assays of human phosphate metabolites by nuclear magnetic resonance. Clin Chem 1989;35:392-5.
- Cistola DP, Robinson MD. Compact NMR relaxometry of human blood and blood components. Trends Analyt Chem 2016;83:53-64.
- Delatour V, Clouet-Foraison N, Gaie-Levrel F, et al. Comparability of lipoprotein particle number concentrations across ES-DMA, NMR, LC-MS/MS, immunonephelometry, and VAP: In search of a candidate reference measurement procedure for apoB and non-HDL-P standardization. Clin Chem 2018;64:1485-95.
- Lee H, Shin TH, Cheon J, et al. Recent developments in magnetic diagnostic systems. Chem Rev 2015;115:10690-724.
- Otvos JD, Shalaurova I, Wolak-Dinsmore J, et al. Glyca: A composite nuclear magnetic resonance biomarker of systemic inflammation. Clin Chem 2015;61:714-23.
- Winsten S. Early adventures with NMR and clinical chemistry. Clin Chem 1994;40:2320.
- Skewis LR, Lebedeva T, Papkov V, et al. T2 magnetic resonance: A diagnostic platform for studying integrated hemostasis in whole blood. Clin Chem 2014;60:1174−82.