 Courtesy of Michael Northrop, Biodesign Institute at Arizona State University
Courtesy of Michael Northrop, Biodesign Institute at Arizona State University
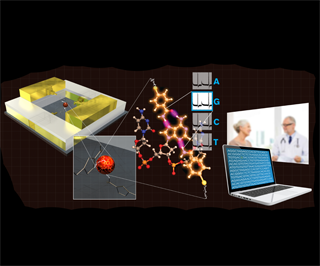
A prototype DNA reader has the potential to make whole-genome profiling commonplace, say the Arizona State University (ASU) Biodesign Institute and IBM T.J. Watson Research Center scientists who developed it. The tiny device—which can distinguish individual chemical bases of DNA when they are pumped past its head—is “thousands of times smaller” than the width of one human hair, according to a press release.
"Our goal is to put cheap, simple and powerful DNA and protein diagnostic devices into every single doctor's office," said Stuart Lindsay, an ASU physics professor and director of Biodesign's Center for Single Molecule Biophysics, in a prepared statement. The authors detailed their methods in the journal ACS Nano.
The device was developed using what they call a “sandwich”—two metal electrodes separated by a two-nanometer insulating layer. “Then a hole is cut through the sandwich: DNA bases inside the hole are read as they pass the gap between the metal layers,” according to the press release.
“The technology we've developed might just be the first big step in building a single-molecule sequencing device based on ordinary computer chip technology," Lindsay explained. “Previous attempts to make tunnel junctions for reading DNA had one electrode facing another across a small gap between the electrodes, and the gaps had to be adjusted by hand. This made it impossible to use computer chip manufacturing methods to make devices.”
In developing this device, the researchers came up with a manufacturing process that allows it to work reliably for hours, “identifying each of the DNA chemical bases while flowing through the two-nanometer gap,” according to the press release. However, they also found considerable variation between devices, so calibration will be necessary to address that issue. Next steps include modifying the device’s technique so that it can read other single molecules, which could prove useful for drug development.