Consider this scenario: A clinician is giving a new monoclonal antibody drug to her patients and wants to measure anti-drug antibodies in serum. There are no Food and Drug Administration (FDA)-approved assays or research-use only kits available for this application, and no reference laboratory offers this test. If you had to develop an assay from scratch, what technique would you choose?
If you were in a clinical laboratory, you might consider making your own enzyme-linked immunosorbent assay (ELISA). But if you were in an academic setting or pharmaceutical company, you might consider as an option surface plasmon resonance (SPR). SPR can be automated, requires few biological reagents for method development, and generates results in just a few minutes. So, if SPR has so many advantages, why aren’t more clinical laboratories using it?
What Is SPR?
SPR is an optical technique that uses refractive index changes at a metal surface to monitor binding interactions in real time and in a label-free manner. In the most common instrument setup (Figure 1), biomolecules such as proteins, antibodies, DNA, or RNA, are immobilized to a metal surface and buffer constantly flows across the surface. Polarized, single wavelength light is directed through a glass prism to the bottom of a metal surface and is reflected back to the detector.
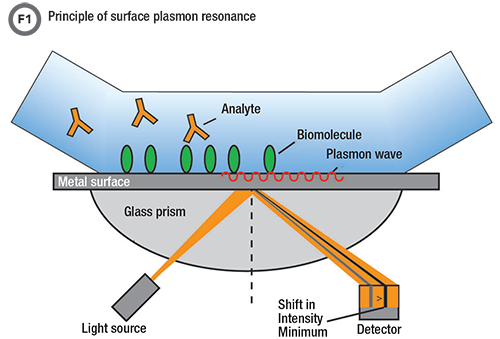
At a certain angle, called the resonance angle, electrons in the metal absorb light resulting in an intensity minimum at the detector. When biomolecules bind to the surface, it causes a shift in the angle at which the light is absorbed. This shift in the intensity minimum is the basis of SPR signal.
The magic happens when, at a very specific angle, called the resonance angle, electrons in the metal absorb the light and cause an intensity minimum at the detector. These electrons, also known as surface plasmons, are very sensitive to the environment at the metal surface. Any changes at the surface affect how the light is absorbed and cause a shift in the angle of the intensity minimum. Thus, when an analyte binds to the immobilized molecule at the surface, a signal change occurs which allows interactions to be measured without labels. The surface can then be washed to remove bound analyte and make binding sites available for subsequent reactions. The data generated from the experiment, called a sensogram, is the SPR signal plotted versus time (Figure 2).
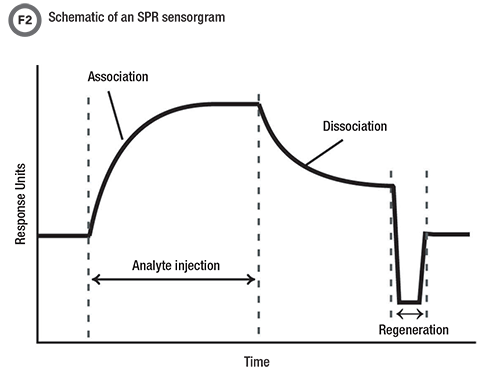
The SPR signal is measured over time to generate a plot called a sensorgram. During an experiment, analyte is injected over the surface. The SPR signal increases as analyte molecules bind to their binding partners which have been immobilized to the surface. Next, buffer is injected and the analyte dissociates. The surface can be washed, or regenerated, and reused in subsequent experiments.
SPR is a very versatile platform in terms of the types of interactions that can be studied and the data they generate. A variety of biomolecules can be immobilized to the surface using different chemistries. Proteins are often covalently immobilized to the surface through amine coupling, while DNA is often covalently immobilized after modification with a thiol group. A variety of molecules also can be attached using streptavidin/biotin.
Although researchers most commonly use SPR to study protein-protein or protein-antibody interactions, SPR has also been used to study DNA-DNA interactions, DNA-protein interactions, and to detect viruses, whole cells, and extracellular vesicles (1,2).
SPR offers several types of data, including kinetic and thermodynamic information about binding (i.e. on- and off-rates, and affinity constants). SPR also can measure analyte concentration or provide qualitative information about whether an analyte interacts with another biomolecule.
Variations of SPR and Similar Biosensors
There are some variations on the common SPR technique described above. For example, SPR imaging (SPRi) monitors SPR signals at specific locations on a surface. Laboratories often use SPRi in multiplexing applications to monitor multiple reaction spots at the same time. Another variation, localized SPR, generates a signal from metal nanoparticles rather than at a flat surface.
Biolayer interferometry (BLI) is also an optical, label-free technique similar to SPR. The physics of signal generation are slightly different, but the measurement process is the same. This technique immobilizes biomolecules to the sensor surface, monitors binding in real time, and can regenerate the sensors. The biggest difference between BLI and SPR is that BLI uses sensor tips rather than a surface: The sensor is dipped into a sample during measurement rather than the sample being flowed over a surface. The advantages of this approach are that there are no fluidics to clog and less buffer is used during experiments.
Applications in Clinical Laboratories
With relative ease, laboratories can develop SPR methods that measure the concentration of protein analytes, and there are many examples in the literature for clinically relevant analytes (2,3). SPR is particularly well suited for measuring antibodies, as an SPR signal is proportional to the amount of mass binding to the surface. Applications relevant to clinical laboratories include measuring the concentration of therapeutic monoclonal antibody drugs (4), detecting the presence of anti-drug antibodies (4,5), and measuring a patient’s antibody response to antigen, often in the setting of autoimmune diseases (6,7). There are also many examples in the literature of SPR methods to measure clinically relevant proteins, including C-reactive protein, prostate-specific antigen, brain natriuretic peptide, cytokines, myoglobin, and troponin (2). However, most of these protein analytes can be measured by FDA-approved, automated immunoassays, and it does not make sense for a clinical lab to develop an SPR method instead of using one of these commercially available assays.
However, SPR can play a role in clinical laboratories when commercial assays are not available, as in the example given at the beginning of this article. Consider developing an ELISA or sandwich immunoassay from scratch—a time-consuming and expensive endeavor because of the need for two antibodies specific to the analyte but directed to different epitopes. One of these antibodies also needs to have a label, which sometimes can interfere with analyte binding or affect the stability of the antibody. If a suitable antibody pair is not commercially available, clinical labs are unlikely to make their own ELISA. Yet, developing an SPR method is much more accessible since the technique requires fewer biological reagents. SPR is attractive in this setting because method development is simplified.
Another advantage of SPR is that it does not require significant sample preparation or sample volume. Typically, laboratories diluted and injected samples directly onto the SPR instrument. Laboratories also can easily multiplex SPR by arranging different surfaces in series or through the use of SPRi, in which biomolecules are immobilized to different regions of the surface. In addition, because signal is generated in real time, results are available much faster compared with ELISA, which is performed in batches and has several incubation and wash steps.
Collecting data in real time also means that SPR provides kinetic information about binding that cannot be determined by ELISA or other equilibrium end-point measurement techniques. This information may have value in diagnostic testing. For instance, the affinity (how much analyte binds at equilibrium) and the on- and off-rates (how fast an analyte binds and dissociates from the immobilized ligand) could serve as signatures of a particular ligand–analyte combination and may be able to distinguish between different analytes.
Investigators have used the additional information generated by SPR to evaluate vaccine efficacy (8), detect biomarkers in autoimmune diseases (6,9), and distinguish between inhibitory and noninhibitory factor VIII antibodies (10), for example. This information may also be helpful in distinguishing interferences from true analyte interactions in commercial immunoassays. However, it is unclear if any of these assays are being used in clinical labs, or if this additional information will have more widespread diagnostic application.
Challenges to Using SPR in Clinical Labs
Clinical laboratories face several barriers in adopting SPR. Analytically, one of the major challenges with SPR is nonspecific binding, which can reduce the sensitivity of the assay. Nonspecific binding is especially important to consider when working with complex biological samples like serum. However, there are several ways to minimize or correct for this. As in commercial immunoassays, labs often add detergents to both the buffer and samples in order to reduce unwanted interactions with the surface. Adding bovine serum to albumin can help “precoat” the surface and prevent proteins in the sample from binding. Finally, labs can dilute samples before measurement.
Labs can also correct for nonspecific binding during the data processing step. Typically, a blank reference surface (a surface without immobilized biomolecules) and a blank sample (a sample without analyte) are used to test for nonspecific binding to both the surface and the immobilized biomolecule. The raw lab can then subtract the raw sensogram observed from these controls from that of the sample to correct for nonspecific binding.
In addition to analytical challenges, clinical labs face several practical challenges in adopting SPR. First, SPR instrumentation is expensive, ranging anywhere from $50,000-$300,000 depending on the throughput or number of channels in the instrument. In addition, only a handful of companies make SPR instruments. Several firms supply SPR and BLI instruments, and these companies primarily design and market their instruments for academic and pharmaceutical laboratories. Vendors’ focus on research and pharmaceutical applications partly explains why clinical laboratorians are not very familiar with SPR and may have the perception that the technique is too complex for their needs. SPR does require some expertise to appropriately design methods and interpret data, as is the case with any laboratory-developed test. Finally, there is currently not a strong need to measure the kinetics of a binding interaction, which is a unique feature of SPR. The other advantages—simplified method development, automation, fast time to result, and others—may also not be crucial, except in limited cases.
Conclusion
At least in the literature, there has been a lot of interest in the use of SPR for clinical applications. There are a number of proof-of-principle papers, and several reviews published about the use of SPR for clinically relevant applications (2,3). But so far, advantages of SPR such as kinetic information, automation, fewer biological reagents, simplified method development, and fast time-to-result have not been sufficient driving forces for widespread adoption, and the use of SPR is still rare in clinical laboratories.
SPR does, however, play a role, particularly for measuring antibodies and analytes that do not have commercially available assays. Will we see it being used more in the future? Perhaps. But this will require work on the part of clinical laboratorians and scientists to continue to improve the technique and explore how to best use it in a clinical laboratory setting.
Katie Thoren, PhD, DABCC, FADLM is an assistant attending clinical chemist at Memorial Sloan Kettering Cancer Center in New York City. Email: [email protected]
References
- Gool EL, Stojanovic I, Schasfoort RBM, et al. Surface plasmon resonance is an analytically sensitive method for antigen profiling of extracellular vesicles. Clin Chem 2017;63:1633-41.
- Mariani S, Minunni M. Surface plasmon resonance applications in clinical analysis. Anal Bioanal Chem 2014;406:2303-23.
- Thoren KL, Pasi B, Delgado JC, et al. Quantitation of infliximab and detection of antidrug antibodies in serum by use of surface plasmon resonance. J Appl Lab Med 2018;2:725-36.
- Real-Fernández F, Cimaz R, Rossi G, et al. Surface plasmon resonance-based methodology for anti-adalimumab antibody identification and kinetic characterization. Anal Bioanal Chem 2015;407:7477-85.
- Metzger J, Landenberg P von, Kehrel M, et al. Biosensor analysis of β2-glycoprotein I–reactive autoantibodies: Evidence for isotype-specific binding and differentiation of pathogenic from infection-induced antibodies. Clin Chem 2007;53:1137-43.
- Buhl A, Metzger JH, Heegaard NHH, et al. Novel biosensor-based analytic device for the detection of anti-double-stranded DNA antibodies. Clin Chem 2007;53:334-41.
- Dennison SM, Reichartz M, Seaton KE, et al. Qualified biolayer interferometry avidity measurements distinguish the heterogeneity of antibody interactions with plasmodium falciparum circumsporozoite protein antigens. J Immunol 2018;201:1315-26.
- Thaler M, Buhl A, Welter H, et al. Biosensor analyses of serum autoantibodies: Application to antiphospholipid syndrome and systemic lupus erythematosus. Anal Bioanal Chem 2009;393:1417-29.
- Kocot C, Schindler AR, Le Blanc A, et al. Biomimetic biosensor to distinguish between inhibitory and non-inhibitory factor VIII antibodies. Anal Bioanal Chem 2015;407:5685-93.