A guiding principle of clinical laboratories is to provide patient test results as quickly as possible while maintaining accuracy and quality. Therefore, advancements in analytical technology that not only improve accuracy and quality, but also decrease the length of time from sample collection to results are highly desirable. In the clinical microbiology laboratory where many traditional culture-based methods take days to yield results, providing diagnostic information in a matter of hours using mass spectrometry (MS) and molecular techniques holds the potential for vast improvements in patient care.
One of the major drivers today for providing rapid microbiology test results is the practice of antimicrobial stewardship, which aims to reduce the amount of unnecessary antibiotics administered before laboratories definitively identify the type of bacterial infection (1). Combining laboratory and pharmacy practices in a stewardship program has several advantages, including minimizing patient exposure to IV antibiotics, decreasing the possibility of microorganisms developing antibiotic resistance, and reducing costs associated with prolonged hospital stays.
While clinical laboratories can make a significant contribution toward rapid identification of infectious agents, limitations still exist. The shift toward molecular techniques for bacterial identification, such as DNA sequencing, real-time polymerase chain reaction (PCR) assays, and peptide nucleic acid-fluorescent in situ hybridization (PNA-FISH), has led to improvements in turnaround time. However, the limited number of commercially available assays and the need to have an idea about which organism is suspected before selecting the appropriate kit and reagents make it difficult to apply truly universal bacterial identification regimes.
Here we provide a brief overview of MS technologies useful in identifying bacterial infections. Because these label-free technologies do not require knowledge of a particular genetic sequence, they offer specific advantages for characterizing the genetic or protein material of bacterial organisms directly, as well as minimizing the technical complexity of the workflow.
The Technologies
Two MS technologies are emerging for bacterial identification: PCR electrospray ionization MS (PCR-ESI/MS) of genetic material, and matrix-assisted laser desorption/ionization time of flight MS (MALDI-TOF/MS) of proteins.
PCR-ESI/MS
PCR-ESI/MS uses the mass-to-charge (m/z) ratio of a PCR amplicon to infer its base composition. The method uses primers targeted to conserved regions in the bacterial genome, as well as species-specific regions for accurate identification, and produces PCR products of different length and composition (Figure 1). In order to identify the bacterial species, this information is compared to a database containing base composition data for a wide variety of bacteria (2).
Figure 1
General Scheme of PCR-ESI/MS
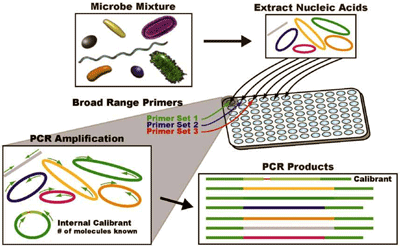 DNA is extracted from the microbial mixture and amplified with broad-spectrum primers. This generates amplicons of varying lengths and masses, which are used to infer base composition and compared to a database of microbial species. Reprinted with permission from Elsevier.
DNA is extracted from the microbial mixture and amplified with broad-spectrum primers. This generates amplicons of varying lengths and masses, which are used to infer base composition and compared to a database of microbial species. Reprinted with permission from Elsevier.
The automated process desalts these PCR products which are then introduced to the MS via electrospray ionization and separated based on mass/charge (m/z) ratio in the TOF mass analyzer. The smaller ions of lower m/z travel faster and reach the instrument’s detector earlier than larger ions of higher m/z. From the spectrum produced, the molecular weight of the amplicon can be calculated. This mass is then used to determine the amplicon’s nucleic acid base composition from a mathematical algorithm that incorporates the combination of masses of the four nucleic acid bases (A, C, T, and G), along with the reverse complement of the DNA strand, to predict the amplicon’s composition. The information from one primer set is combined in parallel with several other loci so that a special software program can compare it to a database of known base compositions and determine the identity of the organism. The laboratory reports the final result with a confidence score, which accounts for the rarity of the amplified region and the number of primers used for identification.
PCR-ESI/MS analysis is fundamentally different from standard PCR assays that require primer and probe selection, making it advantageous for clinical bacterial identification. This assay’s primers broadly amplify microbial DNA, rather than being dependent on knowledge of a particular sequence. Additionally, because PCR-ESI/MS is a direct detection method, multiple amplicons can be identified simultaneously, provided their molecular weights are not identical. One commercial platform is currently in development, the Abbott PLEX-ID system, which offers a wide array of assays, depending on the needs of the laboratory.
MALDI-TOF/MS
MALDI-TOF/MS also employs MS as the detection step, but it is very different from PCR-ESI/MS. In this technology, the molecular weights of proteins and peptides are used to identify microorganisms. The technologist deposits intact microbes onto a sample plate, either from single colonies or liquid culture media, and mixes them with an ionizing matrix (3,4). The sample is then irradiated with a UV laser to ionize the soluble proteins, which are characterized by their m/z using TOF/MS, similar to PCR-ESI/MS. In contrast, MALDI-TOF/MS compares the entire mass spectrum of the proteins to a database of spectra collected from known organisms, and the software program identifies the bacteria based on the best match to spectra in this database. Dominant spectral peaks from highly conserved proteins are stable under a variety of growth conditions, avoiding spectral bias to one particular culture method and assuring reproducibility. An algorithm determines which organism is the closest match to the sample organism, and the laboratory reports identification using a confidence value (Figure 2) (5).
Figure 2
General Scheme of MALDI-TOF/MS Identification of Bacteria
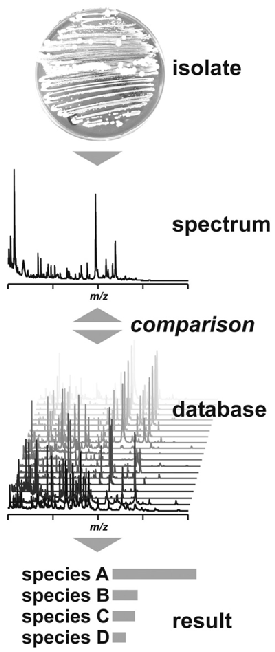 Spectra are generated directly from organisms and are compared to a reference database of spectra from known organisms. Results are reported based on the confidence of the spectral match.
Reprinted with permission from Elsevier.
Spectra are generated directly from organisms and are compared to a reference database of spectra from known organisms. Results are reported based on the confidence of the spectral match.
Reprinted with permission from Elsevier.
Bacterial identification by MALDI-TOF/MS is fundamentally different from biochemical and antigenic methods. The method does not require specific primers, so all microorganisms can be identified using a single assay. This approach offers several advantages, including being broadly applicable to all microorganisms, reducing the number of procedures and reagents required for identification, and requiring less specialized knowledge about microbiology.
Two major platforms offer this technology: the Bruker MALDI Biotyper and the Shimadzu AXIMA Microorganism Identification System, available from bioMerieux as the Vitek MS. While these platforms are very similar in their instrumentation, their scoring algorithm and signal processing differ (6).
Advantages of Rapid Bacterial Identification
Because clinical microbiology laboratories have only used these MS-based technologies for a short time to identify bacteria, no formal studies of their benefits exist. However, many studies have demonstrated the impact of antimicrobial stewardship on patient outcomes. In one, a minor adjustment in the laboratory workflow-—verifying results and releasing them twice instead of once a day—improved turnaround time by 5 hours (7). This change lead to a 1.7% decrease in the mortality rate, a 2-day drop in average length of hospital stay, and a $1,750 average savings in variable costs directly associated with patient care. In a similar study, advances in reporting antimicrobial susceptibility resulted in a 1.4% decrease in the mortality rate, a 1.2-day decline in average length-of-stay, and a $1,466 average total cost savings per patient, amounting to approximately $3 million annually (8). More recently, an analysis found that providing Gram stain results faster significantly reduced mortality rates (Table 1) (9).
|
Table 1
Improvements Afforded by Rapid Diagnostic Methods
|
| |
Mortality |
Length-of-Stay |
Variable Cost |
Total Cost |
| Workflow Improvement (7) |
↓ 1.7%* |
2.0 d |
$1,750 |
$2,395 |
| AST Reporting (8) |
↓ 1.4% |
1.2 d |
$2,812 |
$1,466 |
| Gram Stain Results (9) |
↓ 9.1% |
0.5 d* |
$182* |
NR |
| PNA-FISH Reporting (10) |
NR |
2.0 d |
$1,301 |
$4,005 |
| PNA-FISH Reporting (11) |
↓ 8.9% |
2.0 d |
NR |
$19,441 |
|
*Not statistically significant difference from control group
Abbreviations: NR, not reported; AST, antibiotic susceptibility testing; PNA-FISH, peptide-nucleic acid fluorescent in situ hybridization.
|
Studies demonstrating the benefits of using molecular testing to identify bacteria have also been published, including several involving PNA-FISH. One investigation showed that early results from PNA-FISH significantly impacted length of stay and hospital costs when it came to differentiating potentially infectious staphylococci from contamination due to coagulase-negative staphylococci (10). The authors reported a 2-day length-of-stay reduction, a $1,301 per patient decline in variable costs, and a $4,005 per-patient drop in total costs. A similar study of PNA-FISH method in identifying staphylococci also demonstrated significant benefits, including reducing mortality by 8.9%, length-of-stay by 2 days, and cost of hospitalization by $19,441 per patient.
Although these studies are not specific to MS, they illustrate the potential impact of rapid, broad-spectrum bacterial identification. MS technology has evolved immensely in recent years with the availability of highly automated instruments that minimize the amount of specialized training required. For example, the PCR-ESI/MS platform has a DNA extraction and amplification step on the front end of the analysis that requires hands-on time, but the results are available in as little as 4–6 hours from positive culture bottles. MALDI-TOF/MS platforms also feature workflows with minimal sample handling. The technician applies a colony of organisms, or approximately 1 x 105 organisms from liquid culture, directly onto a MALDI plate for analysis. The remainder of the analysis is automated and takes only 15 minutes.
Advantages of MS Technologies
Studies have demonstrated that the diagnostic accuracy of PCR-ESI/MS and MALDI-TOF/MS are quite high, with positive and negative agreement >94% for both genus and species identification. Both methods also were statistically equivalent (12,13); therefore, the pros and cons of each depend upon the application.
PCR-ESI/MS’s utility in identifying microorganisms is well documented, but this method offers an additional advantage for epidemiological studies. The microbial database is composed of sequences from different species, as well as different strains within those species. Additionally, due to the high specificity of MS, the PCR step allows for partial mismatch of amplification of regions that are similar to the primer binding sequence. This identifies new strains or silent mutations that are relevant for epidemiological studies. PCR-ESI/MS, though more expensive in both startup and variable costs to the hospital, also provides very specific details that can be useful in tracking disease transmission. Furthermore, this technology has the potential to minimize culturing requirements and enable analysis directly from biological materials.
MALDI-TOF/MS also has unique advantages. It is highly automated and can provide results in as little as 15 minutes after >1 x 105 organisms have been grown, whether from subculture or liquid broth. Outside of startup costs, MALDI-TOF/MS is very inexpensive, requiring only the purchase of disposable target plates and matrix. Furthermore, it is user-friendly and requires little training to run. Its simplicity also makes this platform attractive for clinical work, as its workflow can be applied without modification to a variety of microorganisms.
Limitations of MS Technologies
The major barrier to implementing MS technology in today’s clinical laboratories is the capital investment cost and instrument maintenance costs. Both PCR-ESI/MS and MALDI-TOF/MS require a dedicated MS, which can be expensive. Owing to the database composition and development costs, PCR-ESI/MS platforms can be approximately 2–3 times more expensive than MALDI-TOF/MS platforms (14). As is true for most emerging technologies, these methods will be limited to larger reference laboratories until the cost-benefit is fully evaluated or until manufacturers develop smaller, more affordable benchtop options.
Both technologies have limitations, too. A key disadvantage of PCR-ESI/MS is the open platform, which makes ambient contamination with amplicons possible. Therefore, maintaining unidirectional workflow is essential in preventing contamination of PCR reaction mixtures. One of the limitations of MALDI-TOF/MS is the number of organisms required for identification. Depending on the organism, culturing 1 x 105 cells can be a major time limitation and does not eliminate the need for a primary culture. It does, however, shorten the turnaround time by reducing the need for lengthy sub-cultures.
Ready for Widespread Use?
Despite the advantages of MS as a rapid technique for identifying microorganisms, the current state of the technology will not completely replace culture-based testing in the near future. Clearly, MS reduces a portion of the laboratory’s workload; however, a strong need remains for culture-based testing to assess antimicrobial susceptibility. Moreover, culture-based testing may still be advantageous for identifying mutated strains that may not be detected by these molecular techniques. Currently, the cost of implementing these emerging technologies and the familiarity and track record of culture-based testing creates high barriers for laboratories to clear. Nevertheless, MS platforms’ robust analytical sensitivity, accuracy, reproducibility, and broad applicability when it comes to identifying bacteria have the potential to change the practice of clinical microbiology.
While there is still a great amount of work required to realize the benefits of such technology, the future is in sight.
REFERENCES
- Paxton A. Antibiotic program supersizes impact of molecular testing. CAP Today 2011;25:49–52.
- Hofstadler SA, Sampath R, Blyn LB, Eshoo MW et al. The universal biosensor. Int J Mass Spectrom 2005;242:23–41.
- Maier T, Kostrzewa M. Fast and reliable maldi-tof ms-based microorganism identification. Chim Oggi 2007;25:68–71.
- Cherkaoui A, Hibbs J, Emonet S, Tangomo M, et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol 2010;48:1169–75.
- Welker M, Moore ERB. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol 2011;34:2–11.
- Emonet S, Shah HN, Cherkaoui A, Schrenzel J. Application and use of various mass spectrometry methods in clinical microbiology. Clin Microbiol Infect 2010;16:1604–13.
- Barenfanger J, Drake C, Kacich G. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol 1999;37:1415–8.
- Barenfanger J, Short MA, Groesch AA. Improved antimicrobial interventions have benefits. J Clin Microbiol 2001;39:2823–8.
- Barenfanger J, Graham DR, Kolluri L, Sangwan G, et al. CD. Decreased mortality associated with prompt gram staining of blood cultures. Am J Clin Pathol 2008;130:870–6.
- Forrest GN, Mehta S, Weekes E, Lincalis DP, et al. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother 2006;58:154–8.
- Ly T, Gulia J, Pyrgos V, Waga M, et al. Impact upon clinical outcomes of translation of pna fish-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther Clin Risk Manag 2008;4:637–40.
- Kaleta EJ, Clark AE, Cherkaoui A, Wysocki VH, et al. Comparative analysis of pcr-electrospray ionization/mass spectrometry (ms) and maldi-tof/ms for the identification of bacteria and yeast from positive blood culture bottles. Clin Chem 2011;57:1057–67.
- Kaleta EJ, Clark AE, Johnson DR, Gamage DC, et al. Use of pcr coupled with electrospray ionization mass spectrometry for rapid identification of bacterial and yeast bloodstream pathogens from blood culture bottles. J Clin Microbiol 2011;49:345–53.
- Wolk DM, Dunne WM. New technologies in clinical microbiology. J Clin Microbiol 2011;49:S62–S67.

Erin J. Kaleta, PhD, is a clinical chemistry fellow in the Department of Laboratory Medicine and Pathology at the Mayo Clinic in Rochester, Minn.
Email

Donna M. Wolk, PhD, D(ABMM) is an associate research professor and division chief for clinical and microbiology in the Department of Pathology at the Arizona Health Science Center and University Medical Center at the University of Arizona.
Email
Disclosures: Donna Wolk declares that she has received grant/research support and honorarium expenses from bioMerieux, Abbott Molecular, and Ibis Biosciences.