Genotyping a few specific sites of the virus has emerged as an effective method. Several approaches use panels of SNP specific primers/ probes. I have developed a fragment analysis approach to detect COVID-19 variant by capillary electrophoresis called CoVarScan. Fragment analysis allows detection of specific deletion/ insertion sizes, common mutational hotspot regions. CoVarScan targets these hotspots in the spike and ORF genes to create a unique mutation signature for rapid, adaptable variant classification.
Omicron changed the game in variant tracking, not only because Omicron has massively increased infectivity, but also all monoclonal antibody therapies but one (sotrovimab) were ineffective. As sotrovimab had limited availability, knowing variant status could dictate therapy. Therefore, CoVarScan was validated in accordance with CLIA specifications in a clinical lab on over 2,000 specimens (sensitivity: 95%, specificity: 98%). The clinical utility was to provide rapid results for patients needing monoclonal antibody therapy.
A major question about variant detection is whether a test can detect the next variant. For most, constant adaptation has been a challenge. This has not been the case for CoVarScan, and even this past week, we found three cases consistent with the “stealth” omicron variant. The official name is BA.2 but is called “stealth” omicron because it lacks the S:Del69_70 mutation responsible for S-gene target failure (SGTF in the TaqPath COVID-19 assay). S-gene target failure is used as a rapid surrogate test to track the rise of omicron variants.
The BA.2 variant is starting to rise in several locations (Denmark, Philippines, Singapore), has increased transmissibility compared to the original omicron strain (BA.1), and is resistant to the monoclonal antibody sotrovimab. Multiple sub-lineages are not unusual; the Delta variant (B.1.617.2) was one of 3 sub-lineages arising from India at the same time. However, given differing spike protein mutations, BA.2 could have alternative monoclonal antibody therapeutic options.
We have identified seven BA.2 variants and 100% were concordant with genome sequencing results. with the distinct mutational signature described in Table 1 and shown in Figure 1.
|
Omicron (B.1.1.529/ BA.1)
|
Omicron (BA.2)
|
|
Mutation
|
CoVarScan Result
|
Mutation
|
CoVarScan Result
|
|
ORF1A:Del 3675_3677
|
ORF1A: 9bp del
|
|
ORF1A: 9bp del
|
|
N:Del 31_33
|
ORF8: 9bp del
|
N:Del 31_33
|
ORF8: 9bp del
|
|
S:Del 69_70
|
RDR1: 6bp del
|
None
|
RDR1: WT
|
|
S:Del 143_145
|
RDR2: 9bp del
|
None
|
RDR2: WT
|
|
S:Del 211 +
S:Ins 214EPE
|
RDR3-4: 6bp ins
|
None
|
RDR3-4: WT
|
|
S:L446
|
L452: drop
|
None
|
L452: WT
|
Table 1. Mutational differences between BA.1 and BA.2
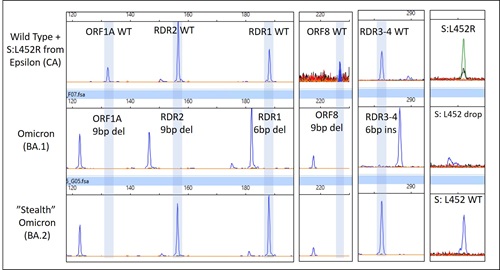
Figure 1. Capillary electrophoresis result of mutations in selected targets of CoVarScan. A wild type variant is shown on top for reference, Omicron (BA.1) in middle panel, and BA.2’s mutational signature is visible in the bottom panel.
Lastly, I think we all realize that this Omicron is not going to be the last COVID-19 variant we encounter. A common critique of SNP based variant genotyping assays is that they require adjustment for new variants. However, this method has required no design adjustment since pre-Delta and has identified unique mutational signatures for each variant. I would go out on a limb to say it will continue differentiating future variants.