In May 2013, an 18-year-old patient arrived at the University of California-San Francisco emergency department in status epilepticus after reportedly taking two blotter papers of lysergic acid diethylamide (LSD). Over the course of his hospital stay, clinicians ruled out other causes of seizures and suspected that his episode was related to the ingestion. However, LSD does not typically cause seizures, and routine drug screening didn’t provide any clues. Extended toxicology screening was requested, and using liquid chromatography-high resolution mass spectrometry (LC-HRMS), we identified dimethoxychloroamphetamine (DOC) in the patient’s urine and serum.1 DOC is a potent hallucinogen that is often mistaken for LSD, and at high doses, has been reported to cause seizures.2
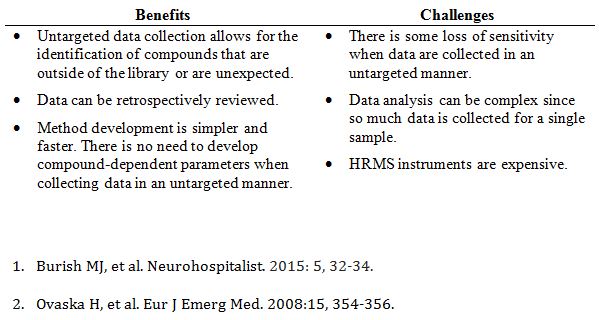
The DOC case is a great illustration of why HRMS is an attractive option in the setting of broad-spectrum drug screening. High-resolution instruments are usually operated in an untargeted manner, which means that accurate mass information is collected on every ion that reaches the detector. This is advantageous for a few reasons, but most importantly, it allows for the identification of compounds that you don’t expect, as was the case with DOC. In contrast, our existing method, which uses a nominal mass quadrupole linear ion trap instrument, collects targeted data on a predefined set of compounds. At the time of the patient’s presentation, our method did not include DOC, so we would have been unable to identify it in the patient’s samples, potentially resulting in a longer hospital stay and more follow-up tests.
While much of the interest in LC-HRMS revolves around identifying unexpected compounds, the vast majority of samples will contain common drugs that are in your library. So, we wanted to know - how well does LC-HRMS work in routine situations? How well does HRMS identify drugs and metabolites when using library searching? And, importantly, how does this compare to nominal mass instruments that collect data in a targeted manner?
In order to address these questions, we developed a broad spectrum drug screen on our LC-MS/MS (ABSciex 3200 QTRAP) and LC-HRMS (ABSciex 5600 QTOF). Both methods were identical in terms of sample preparation, LC method and data analysis software. The LC-HRMS collected untargeted data (full scan positive ion mode with information dependent acquisition of product ion spectra); and the LC-MS/MS collected targeted data (scheduled MRM positive ion mode with information dependent acquisition of enhanced product ion spectra). To compare the ability of each instrument to identify drugs and metabolites, we ran 100 routine clinical urine samples on both methods. We analyzed each dataset for 200 drugs using a spectral library built in-house. We also ran these samples on a third mass spectrometer and used patient prescription information, immunoassay and GC-MS data in order to confirm discordant results.
Overall, we found that the LC-HRMS method performed similarly to the LC-MS/MS method; about 90% of the LC-HRMS results matched the LC-MS/MS results. LC-HRMS found 523 positive hits in the patient samples, of which, 509 (97%) were verified by another method. This means that only 14 hits were deemed to be false positives. The LC-MS/MS method identified 584 positive hits, of which, 543 (93%) were verified by another method. In general, the LC-MS/MS was slightly more sensitive than the LC-HRMS method, but we also observed more false positives.
So, is LC-HRMS better at identifying common drugs? The answer is yes and no. Yes, because you can identify compounds with high confidence and have the ability to detect unknown or unexpected compounds. No, because you do
lose some sensitivity when collecting data in an untargeted manner.
LC-HRMS is certainly a powerful technology, but we are just getting our feet wet. As with any new technology, there are challenges with implementing LC-HRMS in the clinical laboratory. Our goal is to figure out how to overcome these challenges and how to best use LC-HRMS.
What do you think? Does LC-HRMS have a place in the clinical laboratory (now or in the future)?
Benefits and challenges of LC-HRMS (compared to targeted LC-MS/MS methods)