What is commonly known as next-generation genetic sequencing (NGS) made its debut just 7 years ago with the first published report of a commercially available platform being used to dramatically boost throughput in comparison to existing methods. Since then, the scientific literature has been brimming with NGS-related studies, and the technology is very rapidly on its way to replacing the current molecular diagnostic gold standard, Sanger sequencing, a technology developed in the 1970s. The rate of advancement has been so fast the medical community is in some ways struggling to keep pace, and numerous infrastructure and policy issues—many of them lab-related—need to be sorted out before NGS is ready for routine clinical use, experts caution.
"The technology is there. That doesn't mean the ability to exploit the technology most effectively is there. We have lots of hurdles before we can readily implement it in the clinical realm," said Jeff Boyd, PhD, senior vice president of molecular medicine at Fox Chase Cancer Center in Philadelphia. "In the coming years, you'll slowly see major institutions bringing these next-generation sequencing technologies to bear. However, if they're not prepared with multidisciplinary soup-to-nuts teams, in-house, onsite, it'll be rough sledding indeed in terms of getting a physician-friendly report that makes a meaningful difference in a patient's life."

New technologies for DNA sequencing stand poised to enter the clinical arena.
But a major hurdle to adoption will be analyzing the voluminous data output
and interpreting it for clinical decisions.
Higher Throughput, Lower Costs
With the field still in its infancy, even the definition of NGS is up for debate. Although most researchers think of it as massively parallel sequencing, others shy away from that description. "Massively parallel sequencing is kind of a misnomer that doesn't quite capture the concept. I just think of next-generation sequencing as the alternative to Sanger sequencing," said Michael Metzker, PhD, associate professor of molecular and human genetics at Baylor College of Medicine and assistant director of future technologies at Baylor's Human Genome Sequencing Center in Houston. "It's a complicated mess of different techniques trying to sequence DNA faster, cheaper, and at least as accurately as Sanger."
The clutch of commercially available NGS platforms use methods ranging from pyrosequencing and sequencing by ligation to reversible dye termination and real-time single molecule sequencing (See Table, below). They have in common the ability to read exponentially more DNA base pairs per sequencing run than mature methods like Sanger sequencing, which involves copying a DNA template using nucleic acid bases with stop signals. This breakthrough has dramatically pushed down the cost of sequencing, while also making it possible to interrogate much larger regions of the genome—indeed entire genomes—much more rapidly than ever before.
|
Next-Generation Sequencing Platforms
Next-generation sequencing technologies employ different techniques, but all have in common the ability to sequence more DNA base pairs per sequencing run than mature methods like Sanger sequencing.
|
|
Manufacturer
|
Technique
|
Run Time Per Read
|
Base Length
|
Cost
(in 000s)
|
| Helicos |
Reversible
Terminator |
8 days
|
32
|
$999
|
| Illumina |
Reversible
Terminator |
4–9 days
|
75–100
|
$500–900
|
| Ion Torrent |
Real-time |
<1 day
|
964
|
$50
|
| Roche/454 |
Pyrosequencing |
<1 day
|
330
|
$500–700
|
| SOLiD |
Sequencing
By Ligation |
7–14 days
|
50
|
$600–700
|
| Adapted from Mol Ecol Resour 2011;11:759–69; Nat Rev Genet 2010;11:31–46; Am J Clin Path 2011;136:527–39. |
"Historically your technical option for sequencing multiple genes implicated in a particular disorder was to go gene-by-gene and develop a Sanger sequencing option for each gene. This was time consuming, and ultimately a quite expensive endeavor to develop, implement, and provide as a service. So Sanger has been an option, but an unsatisfying one, as the number of genes of interest has increased," said Karl Voelkerding, MD. "We can now use next-generation sequencing to sequence 30 or 50 genes at a time, or nearly the entire coding complement of an individual by exome sequencing. This can be accomplished in a single analytical run at a technical cost that is far lower than what it would take to sequence 30 genes by Sanger sequencing." Voelkerding is medical director for genomics and bioinformatics at ARUP Laboratories and professor of pathology at the University of Utah in Salt Lake City.
These extraordinary advances are changing the very way researchers think about genetic science. "The high throughput nature of the technology and the ability to derive both quantitative and qualitative information has allowed investigators to pursue experiments that were either technically not possible or so impractical by traditional Sanger sequencing that they were not attempted," said Voelkerding.
Breathtaking Changes
By way of reference, the 13-year Human Genome Project, completed in 2003 at a cost of about $2.7 billion, produced the first draft whole genome sequence. The first reference genome, completed in 2004, cost an estimated $300 million. By 2007, genomic pioneer J. Craig Venter reported sequencing his own genome using a whole genome shotgun approach in tandem with automated Sanger sequencing at a cost of about $70 million. Now, just 5 years later, whole genome sequences can be had in about 2 weeks for less than $5,000, and the days of $1,000 whole genome sequences are in sight. Indeed, earlier this year, two companies, Life Technologies and Oxford Nanopore, announced that their novel platforms featuring rapid and real-time sequencing, respectively, will be available by the end of the year at a cost of $1,000 or less.
The pace of change has been breathtaking, even to seasoned researchers. "The impact of next-generation sequencing is illustrated most starkly by the out-of-pocket expenses for whole genome sequencing versus sequencing, let's say, a five-gene panel. The latter costs about $4–5,000, but now we can order a whole genome for just over $3,000," observed Euan Ashley, MRCP, DPhil. "So the irony of a world where you pay several thousand dollars and get the coding regions of only five genes versus paying slightly more than half that and getting both the coding and non-coding regions of the whole genome is kind of bizarre. I've been involved in a few different areas of science and medicine and nothing has moved this fast. It's stunning." Ashley is assistant professor of cardiovascular medicine at Stanford University, and director of the Stanford Center for Inherited Cardiovascular Disease in Palo Alto, Calif. He led the team that carried out the first clinical analysis of a genome, that of his colleague, Stephen Quake, DPhil.
Jump to Whole Genome Sequencing?
If NGS is outmuscling Sanger sequencing so much, will the clinical adoption of NGS simply leap past gene panels to whole genome sequencing? Not quite yet, according to experts. "That's a really interesting question and one we're debating in my group. From a lab wet bench standpoint it's actually easier to prepare DNA for whole genome sequencing than it is for a multi-gene panel because with multi-gene panels you also have to perform an enrichment or selection procedure to enrich for the genes of interest," Voelkerding explained. "There will be a cost point in the not too distant future wherein the price of whole genome sequencing will further raise the debate. It might be routine to sequence the whole genome and then just look at the regions of interest in relation to interpretation of the signs and symptoms of the patient."
Boyd concurred. "Right now and for the next several years we'll be looking at this technology in the clinic as a panel-based approach interrogating targeted hotspots, amplicons, and the like. Where whole exome sequencing becomes much more interesting is in the research space. No matter how little whole genome sequencing costs and how fast it can be done, it's not as useful in the clinical realm in comparison to targeted custom panels, which are actionable," he said. "Panel-based approaches will improve by virtue of what's discovered in the research setting using whole genome sequencing."
The Data Crunch
A key issue influencing adoption of NGS, whether whole genome or targeted sequencing, is being able to analyze the voluminous data output. "Anybody can go out and buy an instrument. And the protocols of sequencing are fairly well worked out, although it requires a lot of training. However, it's the handling of that information, of analyzing and distilling it, that's still very challenging," said Metzker.
Voelkerding agreed, although he pointed out that similar to mass spectrometry vendors, NGS manufacturers are taking strides to make their instruments easier to operate and the results easier to analyze. "Once you receive the data, going through it all and determining its relevance is a very involved process that requires well-trained, well-educated individuals. Even if it's made much more straightforward by software applications, it doesn't take away the fact that someone has to be asking the questions of what you're looking for in the data. And right now that takes an infrastructure of personnel who can navigate through sophisticated computer programs. I don't see that changing immediately."
A Question of Ethics
Aside from merely interpreting genetic data, a myriad of ethical issues also need to be sorted through before NGS is ready for routine clinical use. If drastic cost reductions make it possible for most every patient to have a whole genome sequence in their electronic health records, does that mean it should be done? What privacy standards should prevail? What should be shared with patients when the complex role of genetics in many diseases still remains to be ironed out?
"With diseases like diabetes, there are a lot of known and still unknown genes involved, and many of them are regulatory and acting in the background. So assigning risks associated with those genes will be very complicated. Not only will it take a long time before genome-based outcomes are used for those areas, but some patients may not want to know this information," said Ryan Kim, PhD, director of DNA technologies and expression analysis cores at the UC Davis Genome Center in Davis, Calif. "There's also some debate about carrier screening, where potentially thousands of people could be involved and have good chances of their offspring being carriers of or having diseases. Yet the systems to deal with all of this are not in place."
Addressing Analytical Issues
Though considerable emphasis rightly has been placed on bioinformatic and ethical concerns, numerous analytical issues also have yet to be resolved. "The technology's been moving so rapidly and it's different enough from the way sequencing's been done using other technologies, that there's been an absence of guidance for clinical labs in implementing next-generation sequencing to assure adequate quality of test results," observed Ira Lubin, PhD, genetics team lead in the Center for Disease Control and Prevention's (CDC) Division of Laboratory Science and Standards.
Lubin and his colleagues at CDC spearheaded a work group charged with addressing quality assurance and quality control issues critical to bringing NGS into clinical practice. The group came up with preliminary recommendations and hopes to publish a guidance document later this year. The CDC-led effort is one of many. For instance, Voelkerding, who served on the CDC work group, also chairs the College of American Pathologists' NGS working group, which, among other things, is developing quality metrics and checklist requirements for NGS. In addition, the U.S. Presidential Commission for the Study of Bioethical Issues is examining ethics surrounding clinical implementation of NGS. CDC's Division of Laboratory Science and Standards also is collaborating with other agencies to develop a DNA reference sequence.
Such initiatives are essential to disseminating NGS, experts agreed. "We shouldn't be talking about moving forward without a full discussion of these issues," said Ashley. "I don't think nearly enough attention has been paid to them. There's been a real excitement—and I've been part of it—that this can be transformative for medicine. However, it's moving so fast we have to be careful not to get so wound up in the excitement that we forget to pay attention to ethical and quality issues. We need to be sure that we're making the right use of the technology and not just using it because it's there."
He and others cited numerous performance issues that need to be sorted through, spanning the gamut from accuracy, precision, and sensitivity/specificity to reportable and reference ranges (see Table, click below). "For the type of sequence variance that your test is targeting, it may be difficult to find a sufficient number of patient samples having disease-associated sequence variants to establish these parameters using traditional methods," explained Amy Gargis, PhD, Oak Ridge Institute for Science and Education fellow in CDC's Division of Laboratory Science and Standards. "However, other variants in similar regions of the genome or of a similar type of mutation may be adequate for you to document your repeatability and reproducibility. That is something that labs are doing right now to get around that problem."
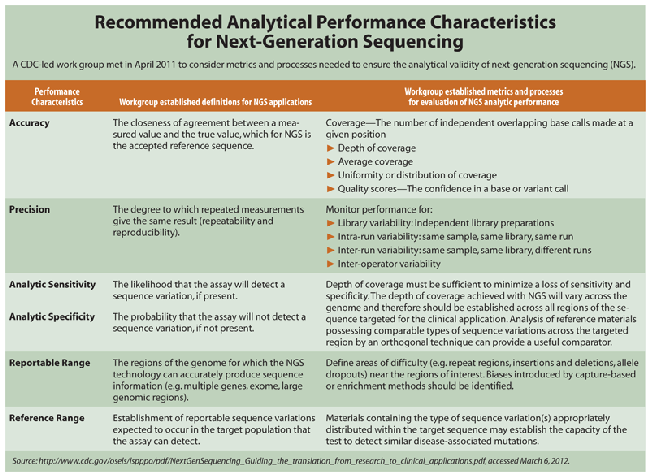
Another example is reportable ranges. "For a traditional lab test you'd say, we can measure the level of this analyte between two specific values. But when you're talking about the whole 3 billion bases of the genome, what do you say?" said Lisa Kalman, PhD, coordinator of the Genetic Testing Reference Materials Coordination Program in CDC's Division of Laboratory Science and Standards. "You can't sequence the whole genome due to technical reasons. Parts of it are just not sequencable because the sequences are very repeated and hard for the technology to deal with. So our working group considered defining reportable range in terms of the regions your test should provide accurate results for."
Given the analytical imprecision still inherent in NGS, many have taken the approach of Ashley and his lab. "I don't think there's a lab yet that would sign off on a clinical genetics test report without validating the call for that position using a different technology," he said. "And I as a clinician would not want to counsel a patient on the basis of a next-generation sequencing result that had not been validated with the current technology at the positions of interest. So what we have is a situation where next-generation sequencing is used like a screening test, if you will. However, the final clinical test result is still signed off by using technology that's currently available."
Also not to be left out of the mix are regulatory and reimbursement issues. "These lab-developed tests appropriately come under Food and Drug Administration oversight, but we're all struggling in a very short period of time to figure this out. Hopefully we can work with the government in rapidly crafting some regulatory strategies that both protect the public and aren't so onerous they prevent the implementation of some very sophisticated, exciting technology," Boyd said. "These tests also aren't cheap, in the sense that even with gene panels, you're still talking about a few thousand dollars. That's a lot of money to many people if it's out-of-pocket. So we're struggling with third party reimbursement and spending a lot of time seeking a path forward that makes sense."
Where Do Clinical Labs Fit In?
With the NGS sands shifting rapidly but not flowing into routine practice just yet, what should clinical laboratorians do? "I don't think anybody should be on the sidelines waiting," cautioned Metzker. The field is moving very quickly and there are many of opportunities for discovery and great science. But one also has to be cautious. One way to do that is through partnerships with groups already experienced with next-generation sequencing. That's a great way of getting your feet wet without being completely isolated and trying to figure out things others already have figured out."
Voelkerding urged laboratorians to learn as much about NGS as possible. "Even if a lab isn't going to be adopting this technology immediately, the director of that lab can be in a situation wherein he or she triages requests for diagnostic analyses," he observed. "So it's important for lab directors nation-wide to begin a self-education process. Since this is all new technology and new data analysis, there are no textbooks one can refer to. It's a self-education process with a steep learning curve that requires reading reviews and attending educational workshops in this area."
By getting up-to-speed themselves, laboratorians can be an important resource for clinicians, many of whom are unprepared for the brave new world of NGS, according to Kim. "The medical community's not ready to deal with that kind of information. When patients start bringing their own gene sequencing data and presenting it to their doctors and asking whether or not there's any significant medical information behind it, the current generation of doctors will not be equipped to answer their questions."
Disclosures: Dr. Ashley is a co-founder of and consultant to Personalis, Inc., a company focused on genomic interpretation.