The virus that causes COVID-19, SARS-CoV-2, is prone to mutate over time, especially within patients who remain chronically infected over months, resulting in genetic variations in the population of circulating viral strains. Variants are defined by an accrual of mutations, some of which are naturally selected because they convey a fitness advantage to the virus, perhaps by such mechanisms as increasing transmission, evading the immune system, or facilitating binding of the virus to its receptor on target cells (creating “Variants of Concern,” or VOCs).
Concern has been building among scientists that new variants will dominate in American infections by Spring, a challenge exacerbated by the inability of the vast majority of nucleic acid amplification tests (“NAAT” which include PCR tests) and antigen tests to discriminate variants versus wild type in SARS-CoV-2 positive samples.
Detecting Variants Quickly With Genomic Surveillance
Genomic surveillance seeks to monitor positivity for variant spread to inform public health decisions. The lack of NGS (Next Generation Sequencing) gene sequencing capacity in the U.S. with which to confirm specific variant identity is creating demand for tools to supplement and strengthen the sequencing capacity of participating laboratories nationally. State Departments of Health now are simultaneously concerned about following the lineage of VOCs and monitoring brand new variant lines as they emerge. Containing variant spread is especially important as we build national immunity via the vaccine rollout. The national NGS bottleneck makes monitoring these dual paths of pathogenesis even more difficult, despite the urgent need to obtain the data.
Schrodinger’s Variant
In the mid 1930’s, Erwin Schrodinger used his famous cat analogy to explain (to those of us more terrestrially bound) a paradox postulated by Einstein that suggests physical objects can be simultaneously in multiple states. The analogy has been amply applied (1) to COVID-19 since infected people may be both infectious and asymptomatic or presymptomatic. Every inter-human contact must be considered an opportunity to infect or be infected. Now the dualities are expanded by the inclusion of both wild type and variant forms of the virus, and the concerns as to whether a variant escapes immune surveillance in those who have been vaccinated or previously infected.
The identification of certain mutations via simple and cost effective NAAT tests can help identify samples that should be further characterized via NGS, can improve the efficient use of this limited resource and can potentially assist with the early detection of new variants in patients, which may help reduce further spread of infection.
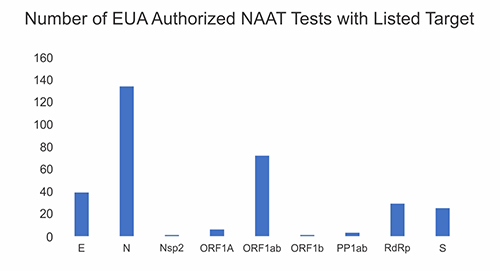
A survey of the EUA (Emergency Use Authorized) nucleic acid amplification tests (NAAT) as of February 25, 2021 revealed that a minority of tests target the S gene that encodes for the Spike protein that mediates binding of SARS-Cov-2 to its receptor to initiate infection. (Figure 1). Of this minority, only the Linea™COVID-19 test targets two amplicons within the S gene. As defined by the EUA, positivity for either amplicon constitutes a positive diagnosis.
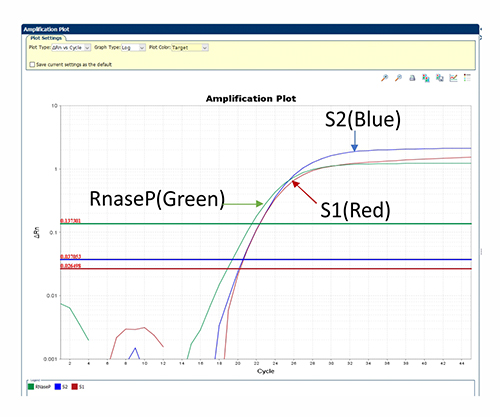
Positive of S1, S2, and Rnase P signals
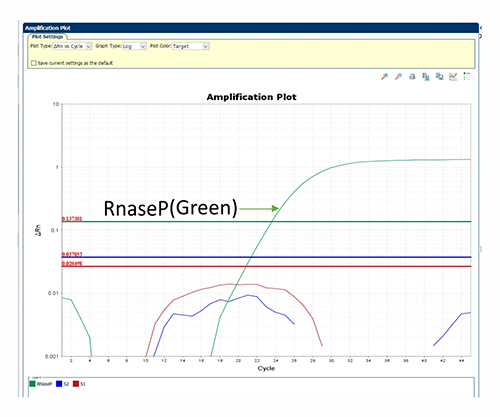
Negative of S1 and S2 signals, Positive of Rnase P signal
Improving the Efficiency of Limited NGS in the US
On January 8, 2021, the U.S. Food and Drug Administration announced in a safety communication that Applied DNA Sciences’ LineaTM COVID-19 Assay Kit was one of a select few NAATs that could potentially identify SARS-CoV-2 variants (including, but not limited to the B.1.1.7 variant) that contain a specific S-gene mutation known as 69-70del. Due to the Linea COVID-19 Assay Kit’s dual S-amplicon design, the overall sensitivity of the Assay Kit is not impacted, and a positive result is still obtained for variant-infected samples.
We have recently detected multiple variants via S-gene dropout from results obtained with the Linea COVID-19 Assay Kit. Multiple laboratories across the US have begun use of the Assay Kit to select for variants from the positive samples identified by variant-agnostic tests. Various state Departments of Health have then identified and confirmed the variants via NGS. We are examining the potential to pool positives to increase the potential throughput for selective genomic surveillance.
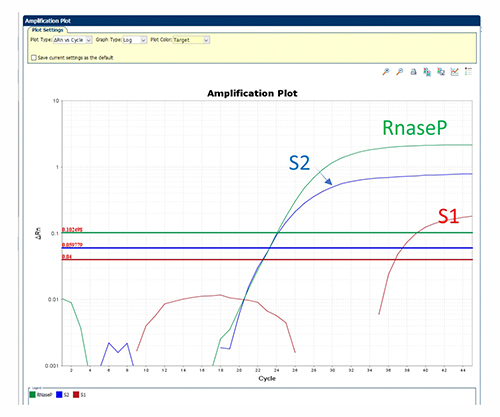
In this variant, S1 signal was significantly reduced. The Ct value for each target is: S1: 36.6, S2: 24.5, RNase P: 25.3
This use case highlights the importance of utilizing a NAAT molecular diagnostic assay that can easily and cost-effectively identify potential variants. We believe that the identification of known VOCs, and new variants from other phylogenetic paths, is essential to containing the spread of COVID-19, even in (and especially in) the face of a successful vaccination campaign. Learn more at the Applied DNA Sciences website.
James A. Hayward, PhD, is the Chairman, President & CEO of Applied DNA Sciences, Inc.
References
1. COVID-19: Leaving lockdown – Of Schrodinger, cats, testing and masks, editorial, George Alexander Thomson, Int J Clin Pract. 2020; 00:e13519