
The Centers for Disease Control and Prevention (CDC) have recommended routine voluntary HIV screening of all Americans between the ages of 13 and 64 years during regular health encounters since 2006 (1). However, CDC data show that fewer than half (43%) of nonelderly adults in the U.S. have ever been tested. Moreover, researchers estimate that 15% of those living with HIV are unaware of their infection and can unknowingly infect others (2). Increased testing, particularly during initial stages when HIV is most infectious, remains an important step in decreasing new cases.
Although the evidence for increased screening is compelling, the approach to using the different types of HIV tests themselves requires expert judgment to serve diverse populations and mitigate the secondary effects of potential false-positive and false-negative results. This article explains how our laboratory developed a machine learning algorithm that allows personalized workflows for HIV screens based on their classification as likely true or false positive, improving our care for our low prevalence and high-risk populations.
MORE CHOICES FOR HIV TESTING
The most analytically sensitive HIV screening tests in regular use in the United States are termed fourth-generation (HIV4G) and fifth-generation (HIV5G) tests. With HIV4G, detection of the HIV p24 antigen was for the first time included with the detection of HIV-1 and HIV-2 antibodies. This reduced the time between infection and positive HIV screen from around 4 weeks to around 2 weeks—critical for detecting HIV during the most infectious period (3).
But HIV4G provides only a single positive/reactive or negative/nonreactive result, requiring follow-up testing to differentiate HIV-1 antibodies from HIV-2 antibodies or HIV-1 p24. CDC recommends that a positive HIV4G be followed by a differentiation test to determine if HIV-1 or HIV-2 antibodies are present. If the differentiation test is negative or otherwise unclear, the algorithm continues to nucleic acid amplification testing (NAAT) for HIV- 1 (4).In HIV5G, a result is provided for each component (HIV-1 antibodies, HIV-2 antibodies, HIV-1 p24 antigen); thus there is potentially less follow-up required.
There continues to be debate around the use of a serologic differentiation assay following HIV4G positive screens because a quantitative viral load NAAT for HIV-1 can provide clinically actionable data that the differentiation assay cannot (5). Additionally, HIV-2 infection in the U.S. is rare and almost exclusively associated with travel in endemic areas (6).
In considering how to best serve our population with HIV5G screening, we recognized that we have several distinct populations to serve. We provide HIV prenatal screening to a large women’s hospital. This is a low-prevalence population in which we expect to see increased false positives. False positives are a concern because in the U.S. and many other countries, HIV is a reportable disease, which means that every positive HIV screen is automatically provided to the department of health. Moreover, for many people a positive HIV screen in their medical record, even if later confirmed as a false positive, is psychologically distressing or culturally unacceptable.
We also serve community clinics and emergency departments that have a higher prevalence and thus more true positives. For these patients, a priority is providing them with results as quickly as possible to prevent unknowing HIV spread.
We asked ourselves if there was a way to predict if a screen was likely a false or a true positive, and then use that information to determine our workflow for the specimen. Retrospective data and some basic machine learning allowed us to answer this question.
IDENTIFYING A PROBLEM THAT COULD BE SOLVED WITH DATA
We realized in our workflow that there were patterns in patient samples that were reactive by HIV5G but subsequently tested negative by NAAT. Often these samples had reactivity for both HIV-1 and HIV-2 (very rare in the U.S.), had low-level reactivity, or were reactive for all analytes. One approach to this problem would have been creating a series of rules for medical laboratory technologists to check, or for us to program into our laboratory information system (LIS). Instead, we decided to explore whether a basic machine learning approach could help us in appropriately classifying reactive results as “likely true positive” or “likely true negative.”
If likely true positive, we could release the result immediately so that the patient could get confirmatory testing via NAAT and be connected to care. If likely false positive, we could assess via orthogonal serologic testing, reach out to the physician to discuss patient history, or result the patient’s screen as indeterminate and in need of follow-up confirmatory testing.
HIV screening as a machine learning problem has the advantage of reasonably easy access to the correct answer about the patient’s diagnosis most of the time. Reactive screens will be followed up with NAAT, thus both the test we are evaluating (HIV5G) and knowledge about whether that test is accurate (NAAT results) are housed within our LIS.
While NAAT still has limitations, we used it as our gold standard for a retrospective evaluation of all HIV5G screens performed in our laboratory between 2016 and 2018. Most patients with a reactive HIV5G screen had NAAT follow-up by our clinical laboratory, but some did not. We had to chart review those cases where NAAT was not available in the database to determine if they were true-positive or false-positive results. Of 60,587 assays, 453 were reactive by HIV5G and 127 were negative by HIV5G but also had HIV NAAT performed (true-negative cases). Of this total of 580 cases that we assessed for HIV final diagnosis, 45 were excluded due to lack of necessary information.
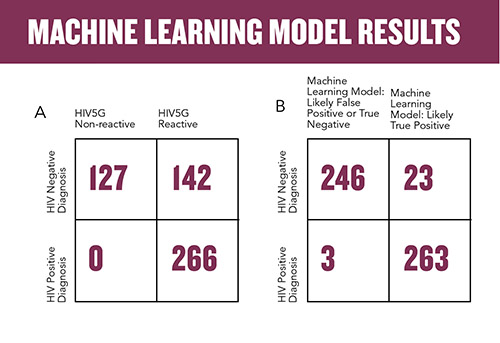
Using the manufacturer’s cutoffs for reactivity, we found that for these 535 cases, the accuracy was 73.5%, with 142 of 408 reactive results being false positive (Figure 1a). It is worth noting that in the calculation of accuracy, we did not include the tens of thousands of presumptive negative cases, as we were specifically interested in reducing false positives.
For our machine learning model, we chose a radial basis function kernel support vector machine (SVM), programmed in MatLab. This type of model learns how to classify the data into two categories: in our case, false positive versus true positive. We used the three results—HIV-1 antibodies, HIV-2 antibodies, and HIV-1 p24 antigen—from the 535 cases as well as a random sampling of 25% of the presumed negative cases. We used the final HIV diagnosis as the ground truth, which in machine learning parlance refers to the correct answer against which the model is trained.
The SVM was able to correctly classify 119 of the 142 prior false positives (Figure 1b). With this classifier, we were able to improve overall accuracy to 95.1% with a false-positive prediction accuracy of 83.8%. We further refined the model to improve this false positive prediction accuracy and were able to raise it to 94%; however, the simple SVM with an accuracy of 83.8% is easier to implement and provides significant improvement for personalizing patient diagnosis.
WHAT TO DO WITH LIKELY TRUE-POSITIVE VERSUS LIKELY FALSE-POSITIVE RESULTS
Our goal with this algorithm was to provide fast results to patients with likely true-positive results so they could immediately be connected to care for follow-up testing, and to take additional steps to assess likely false positive results which could include discussions with the ordering clinician.
Those samples that are classified as likely true positive will be immediately released into the electronic medical record with appended text directing the clinician to the appropriate confirmatory testing. Reactive HIV5G samples that are classified as likely false positive will be tested by an orthogonal HIV4G test; if both are reactive, the original sample result can be released. If the second test is negative, the laboratory director can reach out to the clinician directly to discuss or result the sample as indeterminate with appended text on the appropriate follow-up testing.
In this workflow, every sample that screens reactive for HIV5G will have follow-up testing performed. Looking back at the three cases that tested HIV5G reactive and had a positive HIV diagnosis but the SVM classified as likely false positive, the workflow may have led to a 1-day delay in delivering results due to the change in laboratory workflow.
As with any new process in the clinical laboratory, quality management is paramount. Continued quality monitoring that compares the classification with the NAAT results is important to detect when there may be model drift. This indicates that model retraining will be required. This can be structured as a monthly report that shows the percentage of classification matches to NAAT results with a concordance threshold as the monitor. Retraining can be considered when this monitor is repeatedly failed.
NEW OPPORTUNITIES TO PERSONALIZE PATIENT CARE
HIV continues to be a global issue to which the clinical laboratory can contribute significantly to reducing disease burden through rapid, accurate HIV screening. Machine learning allows us to integrate the stream of information we receive about our population in our daily testing to inform management of future patients. It is also notable that this approach, based solely on the three results produced by the HIV5G screen, allows us to remove some bias that could be created by attempts to assess a patient as high or low risk for HIV, or grouping populations’ risk by where they seek care.
We have more opportunities awaiting us to implement machine learning. Instead of using only the manufacturer’s threshold for reactivity, determined in a pretest probability design to assess analytical sensitivity and specificity, we are able to use all of our patient data over 3 years to inform the likelihood of a current patient result being true positive or false positive. This provides the laboratory with new opportunities to personalize workflows and provide clinicians and patients with the answers they need rather than just a number from our instruments.
Sarah Wheeler, PhD, FADLM, is an associate professor of pathology at the University of Pittsburgh School of Medicine, medical director of clinical chemistry at UPMC Children’s Hospital of Pittsburgh, medical director of the automated testing laboratory at UPMC Mercy Hospital, and associate medical director of clinical immunopathology at UPMC. +Email: [email protected]
REFERENCES
- Branson BM, Handsfield HH, Lampe MA, et al. Revised recommendations for HIV testing of adults, adolescents, and pregnant women in health-care settings. https://www.cdc.gov/mmwr/preview/mmwrhtml/rr5514a1.htm (Accessed February 2023).
- Branson B, Handsfield H, Lampe M, et al. Revised recommendations for HIV screening of pregnant women. MMWR Recomm Rep 2001;50:1-62.
- Guarner J. Human immunodeficiency virus: Diagnostic approach. Semin Diagn Pathol 2017; doi:10.1053/j.semdp.2017.04.008.
- Branson BM, Owen SM, Wesolowski LG, et al. Laboratory testing for the diagnosis of HIV infection: Updated recommendations. CDC Stacks 2014; doi:10.15620/cdc.23447.
- Branson BM. Human immunodeficiency virus diagnostics current recommendations and opportunities for improvement. Infect Dis Clin N Am 2019; doi:10.1016/j.idc.2019.04.001.
- PERUSKI AH, WESOLOWSKI LG, DELANEY KP, ET AL. TRENDS IN hiv-2 DIAGNOSES AND USE OF THE hiv-1/hiv-2 DIFFERENTIATION TEST — UNITED STATES, 2010–2017. MMWR MORBIDITY MORTAL WKLY REP 2020; DOI:10.15585/MMWR.MM6903A2.