
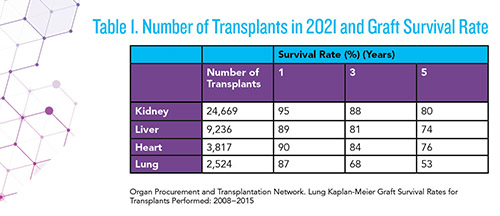
Solid organ transplantation, a modern medical success story, has made tremendous progress in recent decades, aided by imaging techniques, donor-recipient human leukocyte antigen (HLA) matching, and immunosuppressive therapy. In the United States alone, more than 40,000 organ transplants are performed annually, with kidney, liver, heart, and lung being among the most common (1).
Today, major challenges for transplantation are improving long-term graft viability and preserving patient quality of life. Allograft rejection represents the greatest risk of transplant failure among patients. Early identification of subclinical injury, and the subsequent differentiation of injury type, could enable earlier clinician intervention and provide opportunities for personalized treatment. Further, minimally invasive methods of post-transplant monitoring could improve patients’ quality of life while serving as a rejection screening tool and adjunct to histopathology.
Molecular advances in recent decades have led to the emergence of several post-transplant assays that have applications as both screening tools and clinically indicated tests and that address many of the limitations associated with current monitoring practices. These assays have the potential to supplant or complement existing testing paradigms by detecting specific rejection earlier, improving long-term graft viability, and enhancing patient quality of life.
APPROACHES TO POST-TRANSPLANT EVALUATION AND MONITORING
Most clinicians use a systematic approach to evaluate graft health via a combination of monitoring with active surveillance and clinically indicated testing. Active surveillance consists of frequent quantitative testing of late-emerging indicators of injury, including serum creatinine, urinary protein, change in estimated glomerular filtration rate (eGFR), and relevant organ-specific markers (e.g., liver enzymes for liver and pulmonary function for lung). This requires frequent blood draws and a careful balance of immunosuppression adequacy to detect rejection or infection.
For patients presenting with clinical symptoms of injury or suspicion of rejection, clinicians typically order invasive biopsies. Histopathology is the current benchmark for diagnosing allograft dysfunction and identifying the phenotypic causes of rejection. Some transplant centers also rely on serial biopsies to monitor the graft health of patients without clinical indications of rejection.
CLINICAL CHEMISTRY
Conventionally, active kidney transplant monitoring has been conducted using laboratory testing for serum creatinine, eGFR, and total urinary protein because these indicators reflect allograft function. Serum creatinine and proteinuria have been commonly used to predict acute rejection; however, these are non-specific biomarkers that generally signal detectable differences only after allograft injury has occurred. These markers also lack sufficient sensitivity and specificity, and therefore have limited predictive value.
HISTOLOGY
Allograft rejection, as determined by inflammation, is currently best characterized through histological examination. Even though histology serves as the reference standard for diagnosis, serial biopsies to detect subclinical pathologies are painful, risky, and inconvenient for organ transplant recipients. Further, such an approach is cost burdensome on both patients and the healthcare system. Clinically indicated testing will likely lead to biopsy because early diagnosis of acute rejection is important for graft management and improved long-term survival.
Regrettably, clinical interpretation of histology can be subjective and histologic features of antibody-mediated rejection (ABMR) and T cell-mediated rejection (TCMR) may overlap. This ambiguity contributes to high rates of inter- and intra-observer variability among pathologists assessing patient graft status (2). Moreover, the inherent limitations of histology as a reference standard make it difficult to assess the clinical performance of emerging molecular diagnostic techniques, as correlation with histology does not necessarily reflect confirmation of the true disease state. Thus, there are instances where histology alone is insufficient for providing accurate, timely, or conclusive diagnoses.
Molecular-based post-transplant diagnostics are becoming increasingly available and aim to fill the gaps left by traditional histologic assessment of biopsy specimens. Such methods may be ideal for active monitoring because of the ease of sample collection they offer, as well as their reasonable cost, wide availability, and high negative predictive value. Molecular diagnostics could also be useful in clinically indicated cases to offer clarity for graft injury and insight into the pathogenesis of organ rejection.
THE MOLECULAR MICROSCOPE DIAGNOSTIC SYSTEM
The Molecular Microscope Diagnostic System (MMDx) is a microarray-based platform that analyzes expression patterns of messenger RNA (mRNA) from a biopsy core (kidney, lung, heart). It produces a highly informative profile of nearly 1,500 genes known to correlate with both specific rejection (both TCMR and ABMR) and subclinical injury to assess overall allograft health. Results are determined using an ensemble of machine learning algorithms for rejection classification and risk stratification.
Because the technique requires a biopsy, this assay is better suited for clinically indicated testing, as opposed to active surveillance, and could supplement histopathology by distinguishing mechanistic rejection. In previous studies, clinicians agreed that MMDx not only aligned with outcomes more often than histology, but also increased confidence in patient management (3).
MMDx already is commercialized and may be performed in a CLIA-accredited laboratory. Thus, in the presence of other tools such as donor derived-cell free DNA (dd-cfDNA), MMDx could provide essential complementary data to augment clinical decision making. In fact, the correlation of MMDx with dd-cfDNA provides additional evidence that MMDx can enhance diagnostic and clinical certainty compared to histology alone (4). MMDx may also provide insight into instances where elevated dd-cfDNA levels are not accompanied by histopathological evidence of allograft injury.
Less invasive gene expression assays also are available commercially with blood in certain populations. TruGraf, available for liver and kidney, is better tolerated compared to serial biopsy for monitoring and therefore more likely utilized to detect subclinical acute rejection (5). So far, this test has demonstrated a high negative predictive value and may be indicated to rule out rejection, or at minimum, reduce unnecessary serial biopsy.
BIOMARKERS FOR INJURY AND REJECTION
Biomarkers present a minimally invasive opportunity to clarify uncertainties with histological analysis and understand rejection pathogenesis. To that end, they could make a substantial impact on diagnosis and treatment for transplant rejection.
An ideal biomarker should evaluate over/under immunosuppression, rule out rejection, distinguish between ABMR and TCMR, and aid in histological examination. The biomarker should be accessible to both centralized laboratories and transplant centers, and results should be reproducible, regardless of where the test is performed. Additionally, pragmatic concerns such as cost and turnaround time for results would affect clinical applicability and ideally should be optimized for serial use.
DONOR SPECIFIC ANTIBODIES
Post-transplant patients can form anti-HLA antibodies in response to transplant-mismatched antigens. These are commonly referred to as donor-specific antibodies (DSAs). DSAs have become an established biomarker for predicting antibody-mediated rejection, the leading cause of graft loss following a transplant.
At some transplant centers, monitoring DSA is included in the current standard of care and complementary with other clinical chemistry markers. However, ABMR has been known to occur in the absence of DSA, complicating the predictive relationship (6).
CHEMOKINES
Chemokines play an important role in transplant monitoring because they help to mediate the immune response to the transplant. By monitoring the levels of chemokines, such as CXCL9 and CXCL10, clinicians can detect acute rejection and early allograft dysfunction.
Transcriptomic studies of multiple-gene panels have identified candidate combinations that have delivered promising results in predicting acute rejection and risk stratifying patients. CXCL9/10 can be easily measured in sera and urine, and the quantitation of circulating CXCL9/10 can reveal patients’ immune status (7). The assay has been validated to reduce the need for kidney transplant biopsy and, where indicated, leverage biopsy in a more targeted manner.
Importantly, indicators of inflammation are typically not rejection-specific. Chemokines can be used to screen for, but not necessarily confirm, rejection. Nevertheless, CXCL9/10 has potential use as a biomarker to predict rejection severity and a target to customize therapy.
DONOR DERIVED-CELL FREE DNA
Dd-cfDNA has been shown to be a reliable biomarker for allograft injury, and there is evidence that it has the ability to rule out acute rejection based on its high negative predictive value, assuming laboratory turnaround time is reasonable (8). As such, dd-cfDNA analysis is predominately encouraged for clinically indicated testing to avoid biopsy.
Elevations in dd-cfDNA that are not rejection-specific are a chief concern among clinicians, however, and these have the potential to trigger unnecessary biopsies in cases where acute tubular necrosis or opportunistic infections resulting from over-suppression are present. cfDNA also is less emphasized as a surveillance tool because of the assay’s high cost. It is possible that surveillance testing could be implemented in higher-risk populations to avoid particularly invasive heart and lung biopsies.
Another caveat with dd-cfDNA testing is the lack of harmonization with respect to reporting and interpretation. Reporting absolute dd-cfDNA values versus a fractionated ratio may influence the assay’s clinical validity. For example, dd-cfDNA results represented as a proportion can be mischaracterized by changes in recipient DNA that are not related to allograft health, while results reported as an absolute quantification are not influenced by these changes.
Reporting methods must therefore be put into context. A comprehensive algorithm that considers both approaches could be superior. Like MMDx, testing is commercially available, but from a clinical laboratory perspective, practical limitations to adoption include the high cost of the assay, inconvenient blood collections, and false-negative dd-cfDNA results related to poor assay design and reporting.
EXOSOMES
One of the more promising discoveries in recent years has been the use of exosomal mRNA signatures to determine allograft health. Exosomes are categorized broadly as extracellular vesicles that are associated with immune responses. By mediating cell-to-cell communications, exosomal cargo, such as proteins and nucleic acids, could illuminate a diagnostic window into subclinical injury and organ rejection.
Of particular relevance to kidney transplant monitoring are the exosomes excreted in urine, which have high concentrations of mRNA derived from T cells. Urine has long been known as a valuable source of molecules serving as diagnostic markers for renal disease, and urinary exosomes have been shown to be effective screening tools for kidney allograft rejection.
A proprietary exosomal mRNA signature developed by ExosomesDx discriminated between biopsy samples from patients with all-cause rejection and those with no rejection (9). The assay also differentiated acute TCMR from ABMR in kidney transplant recipients, showcasing the ability of exosomal multigene signatures to support more personalized patient management. Given their reliability and stability, urinary exosomal assays could represent a less invasive and burdensome approach to monitor graft health and potentially open the door for at-home collections.
PATHOGEN MONITORING
Transplant recipients are typically dosed for customized target concentrations of immunosuppressants to balance the onset of acute organ rejection (under immunosuppression) against the potential for opportunistic infection (over immunosuppression). For this reason, some transplant centers routinely monitor for cytomegalovirus (CMV), Epstein-Barr virus (EBV) and BK virus (BKV) via PCR-based assays.
For kidney transplant patients, BKV infection is considered the most common viral complication, causing polyomavirus nephropathy (PVN) in up to 10% of kidney transplant recipients. About 50% of PVN-affected patients will experience allograft failure (10).
Adding to diagnostic complexity, infection and rejection share common mechanisms of allograft injury, and the assays used for serial monitoring will likely vary across centers. Pathogens worth monitoring in allograft organs other than the kidney include hepatitis (liver), Toxoplasma gondii and Chagas disease (heart), Pneumocystis (lung), and Aspergillus (all).
MOLECULAR DIAGNOSTICS CAN FILL THE GAPS
There is an urgent need for noninvasive, personalized, precision diagnostics that enable effective monitoring of organ transplant recipients. The gaps left by histological assessment and late-emerging biomarkers of allograft injury provide an ideal opportunity to integrate molecular assays into the transplant diagnostic paradigm with the aim of reducing costs, extending graft survival, and improving patient quality of life.
Although the limitations of the novel assays discussed in this article preclude their adoption as complete replacements for histology, the concomitant use of conventional histopathology, antibody testing, and molecular-based assays appears to offer significant benefits for improving solid organ transplant outcomes.
As molecular approaches gain momentum, machine-learning based algorithms likely will have greater utility than single biomarkers alone, driving the integration of data from various diagnostic platforms and requiring more frequent, interdisciplinary communication between the clinician and the laboratory.
Though greater engagement with government agencies is needed to expand market access and reimbursement, we are on the precipice of a massive shift in the transplant diagnostic paradigm. This will be exciting to witness because it has the potential to augment care, transform therapy, and improve patient survival.
REFERENCES
- Organ Procurement and Transplantation Network. http://optn.transplant.hrsa.gov (Accessed March 2023).
- Furness PN, Taub N, Assmann KJ, et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol 2003; doi: 10.1097/00000478-200306000-00012
- Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: The INTERCOMEX study. Am J Transplant 2017; doi: 10.1111/ajt.14329
- Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation 2022; doi:10.1097/TP.0000000000003838
- Friedewald JJ, Kurian SM, Heilman RL, et al. Development and clinical validity of a novel blood-based molecular biomarker for subclinical acute rejection following kidney transplant. Am J Transplant 2019; doi:10.1111/ajt.15011
- Senev A, Coemans M, Lerut E, et al. Histological picture of antibody-mediated rejection without donor-specific anti-HLA antibodies: Clinical presentation and implications for outcome. Am J Transplant 2019; doi: 10.1111/ajt.15074
- Handschin J, Wehmeier C, Amico P, et al. Urinary CXCL10 measurement in late renal allograft biopsies predicts outcome even in histologically quiescent patients. Transplant Proc 2021; doi: 10.1016/j.transproceed.2021.07.013
- Xiao H, Gao F, Pang Q, et al. Diagnostic accuracy of donor-derived cell-free DNA in renal-allograft rejection: A meta-analysis. Transplantation 2021; doi: 10.1097/TP.0000000000003443
- El Fekih R, Hurley J, Tadigotla V, et al. Discovery and validation of a urinary exosome mRNA signature for the diagnosis of human kidney transplant rejection. J Am Soc Nephrol 2021; doi:10.1681/ASN.2020060850
- Jamboti JS. BK virus nephropathy in renal transplant recipients. Nephrology (Carlton) 2016; doi:10.1111/nep.12728
Marc J. Rumpler, MS, PhD, DABCC (TC, MD), FADLM, NRCC (CC), DLM(ASCP)SC, is a division director of chemistry, toxicology and newborn screening with the Tennessee Public Health Laboratory. He is a clinical laboratory director and consultant based in Nashville, Tennessee. +Email: [email protected]
Christopher McCloskey, MPH, is director of business development in Thermo Fisher Scientific’s Transplant Diagnostics Division based in West Hills, California. +Email: [email protected]
Christopher Lawrence, BSc, MBBS, LLM, MD(Res), FRCP, is an attending transplant and general nephrologist. He is senior director of medical affairs for the Transplant Diagnostics Business of Thermo Fisher Scientific, West Hills, California. +Email: [email protected]