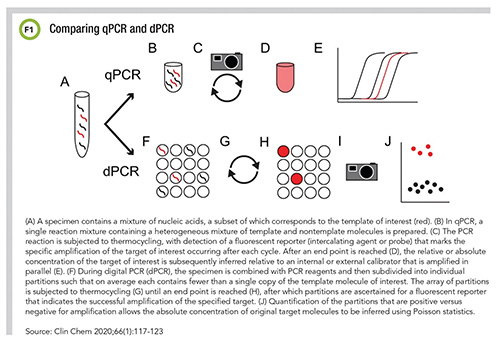
PCR may have entered the general lexicon during the pandemic, but it’s not a new technology. In fact, PCR tests have been used in clinical diagnostics for more than 30 years. Today, PCR technology has been refined to improve its accuracy and workflow. Now real-time PCR (RT-PCR) can be validated not only for detection but quantification, and a new approach called digital PCR (dPCR) even offers direct, absolute quantification of an analyte (Figure 1). So what role will PCR play in the future, both in and out of the COVID-19 spotlight?
CLN spoke to Robin Patel, MD, a professor of medicine, professor of microbiology, the Elizabeth P. and Robert E. Allen Professor of Individualized Medicine, and director of the Infectious Diseases Research Laboratory at the Mayo Clinic in Rochester, Minnesota, about how PCR tests have evolved, how they’re being used right now (with sometimes surprising results)—and how it’s everyone’s job to make sure clinical testing is best serving patients.
PCR testing has been around for more than 30 years. How has accuracy and specificity changed?
At Mayo Clinic, we have been using PCR routinely in microbiology diagnostics since the 1990s. Initially, to detect amplified DNA, we would perform gel electrophoresis followed by a Southern blot. That worked, but was tedious, and interpreting Southern blots can be tricky at times. We performed Southern blot assays to add specificity, and a little sensitivity, but of course optimizing specificity was dependent on probe design. In the early years, there were some laboratories, especially research laboratories, that might have been basing analysis on size of amplified DNA alone, but on the clinical side, we always used Southern blots for specificity.
We transitioned to real-time PCR (RT-PCR) around the turn of the century. That change was advantageous for workflow—it is a lot faster and much less tedious than running a gel and Southern blot. And there is a huge improvement for contamination control.
There are different ways of approaching specificity in an RT-PCR assay. We incorporated probes into our assays to allow for specific detection of amplified DNA rather than generic amplified DNA. We could have used a non-specific dye to detect amplified DNA, but that would not tell us much about whether we had amplified exactly what we were looking for.
In my view, specificity, whether for an old school PCR assay or RT-PCR, comes down to assay design and validation; validation is needed to analytically assess specificity, and is just as important today as it was 3 decades ago.
Test developers need to consider what might cross react in the assay and be falsely detected. From a microbiologist’s point of view, such an ask can be daunting because there are many microbes we haven’t yet characterized, named, or sequenced. On one level, the developer has to think about the disease they are trying to diagnose and on another, the microbes—be they pathogens or non-pathogenic microorganisms—that might be present at the site being sampled.
Interestingly, non-specificity can be helpful, if easily recognized. For example, we’ve had scenarios where we’ve designed assays to detect particular microorganisms but discovered novel microorganisms that hadn’t been known to cause human disease using our assays. We discovered them through the non-specificity of the assays, but we were able to easily recognize what was happening because of our assay design. Since we were careful in how we designed our assays, it was possible to see that there was something being detected that was not what the assays were specifically designed for.
That is obviously not the goal, but it does tell you that we live in a world where we are still learning about—and discovering—microorganisms. Some of them are likely to be pathogens we do not yet understand.
How should clinical laboratories choose between quantitative and qualitative PCR?
This depends on the disease being diagnosed and what the need is for patient care. While quantitative PCR (qPCR) assays are often used for microbial detection, they are typically not validated for quantitative detection. That’s because there are very few microorganisms we have a strong need to quantify and also very few clinically validated quantitative assays. An example of a regularly used quantitative test is HIV viral load measurement; this testing helps guide treatment. Conversely, for many microorganisms, such as Mycobacterium tuberculosis complex, for example, patients are either infected with the organism or not, and having a little bit of that organism is no different, technically speaking, than having a lot of it. It would be like being a little bit pregnant.
In the future, could qPCR assays become more useful in diagnosing other kinds of diseases?
Quantifying accurately and precisely using diagnostic assays in clinical microbiology is harder than people think. Truly quantitative assays usually rely on blood-based specimens because the denominator is relatively straightforward: a volume of blood (or plasma) is standard.
At the Mayo Clinic, many PCR assays we use are performed on non-blood specimen-types, such as body fluids, urine, or tissues. For these specimens, quantifying a denominator is more challenging than the situation for blood, and fraught with variability. Beyond the technical challenge of being able to quantify microorganisms in clinical specimens, the clinical question being answered with such quantification would need definition.
There is also the question of the ideal limit of detection of a qPCR assay. It depends on the microorganism being detected. With microorganisms like Mycobacterium tuberculosis complex, the laboratory needs to have the most sensitive assay possible. But there might be other scenarios where the laboratory is looking for microorganisms that could be part of the normal microbiome. In such scenarios, clinical significance may be based on the relative quantity detected.
It all depends on what the laboratory is trying to accomplish with the assay. One approach is not necessarily better than another for all applications.
Where do you still see opportunities for laboratory medicine professionals to educate clinicians about the nuances of reliability and accuracy of PCR tests?
We need to better understand clinical utility of the assays we have and define and develop the future assays we need. In the end, it is everyone’s responsibility—that is, the laboratory and test-ordering healthcare providers—to make sure our patients receive the appropriate testing they need, and don’t receive inappropriate testing they don’t need.
It is also a shared responsibility that, when patients have testing performed, their results are appropriately reported and acted on. In that sense it is not “education” of clinicians, but the responsibility of the collaborative team to assure the best patient care possible. It is not an us versus them situation.
So, with that context, a simpler answer to your question would be: every day, every minute, all of the time.
Jen A. Miller is a freelance journalist who lives in Audubon, New Jersey. @byJenAMiller