
Costly and invasive tissue biopsies to detect allograft rejection after transplantation have numerous limitations. Assays based on cell-free DNA (cfDNA)—circulating fragments of DNA released from cells, tissues, and organs as they undergo natural cell death—have been intensively studied recently and could ultimately improve our ability to detect rejection, implement earlier changes in management, and even enhance the long-term survival of transplanted organs.
CfDNA assays that circumvent the need for whole-genome sequencing (WGS) and the need for a priori knowledge of donor and/or recipient genotypes have powerful logistical advantages and are currently under clinical scrutiny. In addition, improving knowledge of the organ-specific kinetics of donor-derived cfDNA (dd-cfDNA) following transplantation has also helped optimize these assays. Laboratories also have introduced alternative methods for quantifying dd-cfDNA, such as digital droplet polymerase chain reaction (PCR) and organ-specific DNA methylation patterns. As such, the field of minimally invasive diagnostics based upon cfDNA is increasingly promising, one day potentially replacing traditional tissue biopsies.
The Role of cfDNA in Organ Rejection
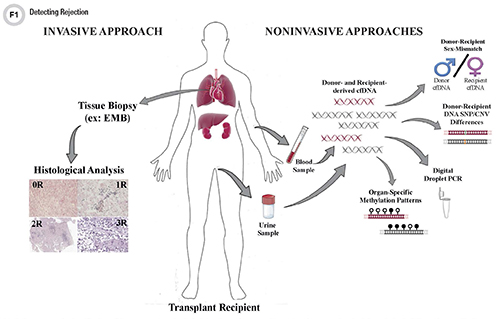
Rejection, referring to injury of a donated organ caused by the recipient’s immune system, can cause allograft dysfunction and even patient death. T-cell mediated acute cellular rejection (ACR) occurs most often within the first 6 months post-transplant (1). ACR involves accumulation of CD4+ and CD8+ T-cells in the interstitial space of the allograft as the recipient’s immune system recognizes antigens on the donated organ as foreign. These T-cells initiate an immune cascade that ultimately leads to programmed cell death (apoptosis) of the targeted cells. As these cells die, genomic DNA is cleaved and fragments of dd-cfDNA, measuring approximately 140 base pairs (bp) in length, are released to join the pool of recipient cfDNA in the blood and ultimately excreted in the urine (2).
Circulating cfDNA has recently been leveraged as a diagnostic tool to replace invasive biopsies in other areas of medicine, including analyzing fetal DNA fragments within the maternal circulation to identify genetic abnormalities in utero and sequencing circulating DNA released from tumor cells to identify cancer-related mutations. In both these cases as well as in transplantation, high-throughput sequencing that identifies and quantifies DNA sequence differences distinguishes between the two different populations of cfDNA derived from distinct sources (2). Three characteristics of cfDNA make it an excellent noninvasive candidate biomarker to detect rejection after solid organ transplantation: It can be obtained from a simple blood draw, its concentration accurately measured, and its nucleotide sequence easily identified. Using cfDNA as a biomarker for ACR is also advantageous since it is derived from the injured cells of the donated organ and therefore should represent a direct measure of cell death occurring in the allograft. Furthermore, cfDNA maintains all of the genetic features of the original genomic DNA, allowing the genetic material released from the donated organ to be differentiated from the cfDNA derived from cells of the recipient that are undergoing natural apoptosis (3).
Frequent and accurate monitoring of allograft health is essential for transplant recipients’ long-term survival. For heart transplantation (HT), endomyocardial biopsy (EMB) is the current gold standard for detecting ACR (4). However, EMBs are costly with significant limitations, many of which are common to all organ biopsies (5-7). Moreover, the invasive nature of EMBs puts HT patients at risk for complications (6,8,9).
Unfortunately, currently available noninvasive methods including echocardiography or magnetic resonance imaging (MRI) lack sufficient specificity and sensitivity to reliably detect rejection (10-13). Blood-based biomarkers, such as cfDNA, represent a promising alternative that could be readily implemented into clinical practice (14-17).
Kinetics of cfDNA During Quiescence and Rejection
Since cfDNA originates from the naturally occurring process of apoptosis, all individuals have detectable levels of cfDNA in their blood (18). For healthy individuals, the majority of circulating cfDNA comes from hematopoietic cells that have undergone natural death related to cellular turnover. Levels of cfDNA fluctuate for multiple reasons including infection, surgery, trauma, or even exhaustive exercise (2,19). Therefore, developing a cfDNA-based assay to detect rejection requires assessing the expected kinetics of dd-cfDNA release into the recipient’s circulation post-transplant. This consideration is especially important since the release of dd-cfDNA over time post-transplant is organ-specific (20-22).
For example, at 1 day post-HT the average level of dd-cfDNA is 3.8 ± 2.3% (20). However, by 7 days the level of dd-cfDNA has declined rapidly and remains consistently low (<1%). During an episode of acute rejection in the heart, the level of dd-cfDNA was found to increase to 4%–5% from a baseline of about 0.06% observed during quiescence. The kinetics of dd-cfDNA observed in the circulation of HT recipients was similar to that observed after renal transplantation (22).
In contrast, recipients of bilateral lung transplants were found to have an average dd-cfDNA fraction of 26 ± 14% on the first postoperative day. Furthermore, the reduction in dd-cfDNA was characterized by levels of dd-cfDNA that declined rapidly within the first week but then slowed and generally remained at 1%–3% (21). However, similar to heart and kidney transplants during an episode of acute rejection, the level of dd-cfDNA increased significantly, climbing to an average of 14%–15%.
Differences in tissue mass and rates of cellular turnover account for this variability in the levels of dd-cfDNA released early post-transplant and during quiescence. For example, differences in circulating dd-cfDNA levels in quiescent bilateral and single-lung transplants can be explained by the difference in cellular turnover, being 107 vs. 58 cells/second, respectively (21). By contrast, in a quiescent transplanted heart, the cellular turnover rate is only 8 cells/second (21-23). Thus, an understanding of the expected levels of dd-cfDNA associated with a given solid organ is essential to facilitate development of organ-specific assays that detect rejection. Once the kinetics of cfDNA release for a particular organ are understood, several methods exist for quantifying the relative amount of dd-cfDNA.
Strategies to Distinguish Recipient- vs. Donor-Derived cfDNA
Donor-Recipient Sex-Mismatch
For organ transplants in which the donor is male and the recipient is female, laboratories can leverage this sex mismatch to calculate dd-cfDNA levels from within the recipient’s total cfDNA pool (17). Researchers first demonstrated the feasibility of this approach in urine samples taken from female renal transplant recipients who had received a kidney from male donors and when they experienced rejection demonstrated elevated levels of dd-cfDNA in their urine that specifically contained regions found on the Y chromosome (17). Although this approach allows for confident diagnosis of rejection in the allograft, sex-mismatch between the donor and recipient is relatively infrequent and not universally applicable.
Donor-Recipient DNA Sequence Differences
An organ transplant can also be regarded as a genome transplant, as the cells within a transplanted organ contain the genetic information of its donor. As such, the concept of genome transplant dynamics (GTD) relies on the presence of genetic differences between the donor and recipient at a particular locus, which then can be leveraged to identify the origin of the circulating cfDNA (20-24). Ideally, the recipient would be homozygous for a single base (for example, AA) and at the same locus the donor would be homozygous for a different base (for example, GG).
Given the genetic heterogeneity between individuals, tens of thousands of potentially informative loci across the genome can be interrogated using high-throughput sequencing to distinguish dd-cfDNA from recipient cfDNA (20,24). This concept was first illustrated using banked samples from cardiac donors to obtain a priori donor genotypes for each donor-recipient pairing. After extracting and sequencing cfDNA from each recipient, the fraction of donor-specific molecules was determined. In samples taken during or immediately preceding a biopsy-proven rejection event, the proportion of donor-specific single nucleotide polymorphisms (SNPs) was found to have increased from <1% to >3%–4% (24).
This early retrospective study has now been validated prospectively. Adult and pediatric heart and lung transplant recipients were recruited and genotypes for each donor-recipient pair were obtained through WGS with an average of 53,423 informative SNP markers identified (20). Overall, early detection of acute rejection was superior to that of AlloMap, the first Food and Drug Administration-approved non-invasive approach to detecting ACR after HT based on transcriptome analysis (25).
Research also has shown that WGS not only provides information about a graft but also a patient’s virome and overall state of immunosuppression. This represents a potentially great advantage unobtainable by other assays (26-28).
However, WGS faces challenges that could prevent it from being implemented routinely in clinical practice. For example, while a recipient’s genetic information can be easily obtained, this is not always true for a donor. Moreover, WGS is costly, labor intensive, and time-consuming.
An alternative method employs a panel of genotyped polymorphic SNPs identified within the pool of extracted cfDNA thereby eliminating the need for a priori knowledge of a donor’s specific genotype (29). Unlike kidney and liver transplants, which often occur between closely related individuals, the donor-recipient pairs for heart and lung transplants typically are not related. GTD requires genotyping of both the transplant recipient and donor. However, in practice, donor genotype information is often unavailable. Here, we address this issue by developing an algorithm that estimates dd-cfDNA levels in the absence of a donor genotype. Our algorithm predicts heart and lung allograft rejection with an accuracy that is similar to conventional GTD. We furthermore refined the algorithm to handle closely related recipients and donors, a scenario that is common in bone marrow and kidney transplantation. We show that it is possible to estimate dd-cfDNA in bone marrow transplant patients who are unrelated or who are siblings of the donors, using a hidden Markov model. Therefore, algorithms have been developed for heart and lung transplants which assume that the donor’s genotype occurs at the same frequency as the general population. Based on these frequencies and comparison to the known genotype of the recipient, the fraction of dd-cfDNA can be reliably estimated from the total pool of cfDNA isolated from a recipient’s plasma sample.
In the case of lung transplantation, this single-genome model, when compared to the methodology using both donor and recipient genotypes, was found to provide comparable fractions of dd-cfDNA. However, when researchers applied this same algorithm to HT, the estimated levels of dd-cfDNA were not as strongly correlated as in lung transplants. This might be related to the lower absolute amounts of dd-cfDNA present after HT. This is another example of organ-specific cfDNA kinetics that can influence assay results and must be taken into account (30).
In the case of renal transplantation, prospective studies have been conducted to ascertain the utility of dd-cfDNA levels, identified using known donor-specific SNPs, as a viable marker for rejection. In one such study, 384 kidney recipients were recruited from 14 clinical sites to provide blood samples at scheduled intervals and at times of clinically indicated biopsies (31). Overall, the study focused on the correlation between the histology in 107 biopsy specimens from 102 patients and the levels of dd-cfDNA found in matched plasma samples. More specifically, 27 biopsy samples from 27 patients with active rejection were obtained along with 80 biopsy samples from 75 patients without active rejection.
In this study, active rejection included acute antibody-mediated rejection (AMR), chronic AMR, and ACR. The assay used in this study employed a 1% cutoff for the fraction of dd-cfDNA to indicate the presence or absence of active rejection and was found to have 85% specificity (95% CI, 79%–91%) and 59% sensitivity (95% CI, 44%–74%). The sensitivity of this assay was greater for discriminating between active and absent AMR, as the use of a cutoff of 1% dd-cfDNA was found to have an 83% specificity (95% CI, 78%–89%) and 81% sensitivity (95% CI, 67%–100%). Notably, in both cases, the sensitivity declined substantially when the fraction of dd-cfDNA exceeded 3%.
To improve specificity and sensitivity of a non-invasive cfDNA-based assay to detect rejection following renal transplantation, investigators also have surveyed the absolute amount of dd-cfDNA (32-33). By interrogating the absolute amount of dd-cfDNA, one can eliminate the artificial changes in the fraction of dd-cfDNA due to increases in total cfDNA levels caused by non-rejection events, such as infection, trauma, or exercise, potentially creating a more accurate assay.
To investigate this possibility, one study employed 32 informative copy number variants (CNVs) based on population frequencies, as opposed to relative proportions of donor and recipient SNPs at given loci (32). All CNVs not present within a recipient’s genome but present within the extracted cfDNA were therefore assumed to represent dd-cfDNA.
Interestingly, while the specificity and sensitivity improved overall with the use of absolute dd-cfDNA levels, this assay also had a greater capacity to distinguish between the presence and absence of active AMR, as opposed to cases of active ACR. In addition, serum creatinine levels were not sufficient in discriminating between active rejection and quiescence, likely because it is more indicative of glomerular function as opposed to kidney tissue damage (31-33).
Another study explored the absolute levels of dd-cfDNA in kidney transplant recipients related to levels of tacrolimus, an immunosuppressant (33). Here, the researchers found that the absolute amount of dd-cfDNA was substantially higher in patients with lower tacrolimus levels (<8 μg/L) in comparison to those with higher drug levels. These data suggest that dd-cfDNA levels also have the potential to detect allograft injury resulting from inadequate immunosuppression.
Laboratories also have proposed alternatives to WGS. Our group explored targeted sequencing of 124 highly polymorphic (minor allele frequency [MAF] >0.4) SNPs using a commercially available panel, next-generation sequencing, and a novel algorithm (34). This approach significantly reduced the total amount of sequencing required, decreasing costs and assay time, and enabling rapid analysis. However, since this assay relies upon differences in MAF between individuals, it would not be robust for closely related donor–recipient pairs, such as seen in living-related kidney donation. It remains to be validated for detecting moderate or greater rejection events.
Laboratories also have explored using polymorphic SNPs to quantify dd-cfDNA combined with the technology of digital droplet PCR (30,35-37). Using 41 highly polymorphic SNPs, stable kidney and HT recipients showed dd-cfDNA fractions of 2%–3% with stable liver transplant recipients having a level of 7% (35).
Conclusions
The use of a costly and invasive tissue biopsy to detect allograft rejection has significant limitations. As such, a minimally invasive assay that can directly and accurately assess the health of the entire transplanted organ represents a holy grail in solid organ transplantation.
The use of cfDNA after transplantation has shown some initial promise, but further study and validation is required to improve our understanding of both the basic biology of cfDNA as well as its behavior post-transplant. At this time, it is clear that important organ-specific differences exist, and patterns of cfDNA release may also differ depending on the type of rejection event. However, cfDNA represents one of the most promising technologies yet developed to complement or even ultimately replace the tissue biopsy.
Sabrina Pattar, MSc, is a graduate student in the department of biochemistry and molecular biology in the Cumming School of Medicine at the University of Calgary in Calgary, Alberta, Canada.
Steven Greenway, MD, is an assistant professor in the department of biochemistry and molecular biology at the Cumming School of Medicine at the University of Calgary and a pediatric cardiologist at Alberta Children’s Hospital in Calgary, Alberta, Canada. +Email: [email protected]
References
- Cornell LD, Smith RN, Colvin RB. Kidney Transplantation: Mechanisms of Rejection and Acceptance. Annu Rev Pathol Mech Dis 2008;3: 189–220.
- Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci USA 2016; 113:E1826–34.
- Naesens M, Sarwal MM. Molecular diagnostics in transplantation. Nat Rev Nephrol 2010;6:614–28.
- Sakakibara S, Konno, S. Endomyocardial Biopsy. Jpn Heart J 1962;3:537–43.
- Khan T, Selvakumar D, Trivedi S, et al. The value of endomyocardial biopsy in diagnosis and guiding therapy. Pathology 2017;49:750–6.
- McMinn J F, Lang NN, McPhadden A, et al. Biomarkers of acute rejection following cardiac transplantation. Biomark Med 2014;8:815–32.
- Pattar, SK, Greenway, SC. Circulating nucleic acids as biomarkers for allograft injury after solid organ transplantation: current state-of-the-art. Transplant Research and Risk Management 2019;11:17–27.
- Baraldi-Junkins C, Levin HR, Kasper EK, et al. Complications of endomyocardial biopsy in heart transplant patients. J. Heart Lung Transplant 1993;12:63–7.
- Mason JW. Techniques for right and left ventricular endomyocardial biopsy. Am J Cardiol 1978;41: 887–92.
- Kindel SJ, Hsu HH, Hussain T, et al. Multimodality Noninvasive Imaging in the Monitoring of Pediatric Heart Transplantation. J Am Soc Echocardiogr 2017;30:859–70.
- Lunze FI, Colan SD, Gauvreau K, et al. Tissue Doppler imaging for rejection surveillance in pediatric heart transplant recipients. J Heart Lung Transplant 2013;32:1027–33.
- Greenway SC, Dallaire F, Kantor PF, et al. Magnetic resonance imaging of the transplanted pediatric heart as a potential predictor of rejection. World J Transplant 2016;6:751–8.
- Butler CR, Savu A, Bakal JA, et al. Correlation of cardiovascular magnetic resonance imaging findings and endomyocardial biopsy results in patients undergoing screening for heart transplant rejection. J Heart Lung Transplant 2015;34:643–50.
- Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS 2010;5:463–6.
- Huo Q, Zhou M, Cooper DKC, et al. Circulating miRNA or circulating DNA-Potential biomarkers for organ transplant rejection. Xenotransplantation 2018;e12444.
- Duong Van Huyen JP, Tible M, Gay A,et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J 2014;35:3194–202.
- Sigdel TK, Vitalone MJ, Tran TQ, et al. A Rapid Noninvasive Assay for the Detection of Renal Transplant Injury. Transplantation 2013;96:97–101.
- Yamani, M. H. & Taylor, D. O. Heart Transplantation. In Current Clinical Medicine 180–186 (Elsevier, 2010). doi:10.1016/B978-1-4160-6643-9.00027-8.
- Atamaniuk J, Vidotto C, Tschan H, et al. Increased Concentrations of Cell-Free Plasma DNA after Exhaustive Exercise. Clin Chem 2004;50:1668–70.
- De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014;6:241ra77.
- De Vlaminck I, Martin L, Kertesz M, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015;2015;112:13336–41.
- Gielis EM, Beirnaert C, Dendooven A, et al. Plasma donor-derived cell-free DNA kinetics after kidney transplantation using a single tube multiplex PCR assay. PLoS One 2018;13:e0208207.
- Bergmann O, Bhardwaj RD, Bernard S, et al. Evidence for cardiomyocyte renewal in humans. Science 2009;324:98–102.
- Snyder TM, Khush KK, Valantine HA, et al. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011;108:6229–34.
- Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene-Expression Profiling for Rejection Surveillance after Cardiac Transplantation. N Engl J Med 2010;. 362:1890–900 (2010).
- Lorio MA, Rosa R, Suarez JF, et al. Influence of immune activation on the risk of allograft rejection in human immunodeficiency virus-infected kidney transplant recipients. Transpl Immunol 2016;38:40–3.
- Simonetta F, Pradier A, Masouridi-Levrat S, et al. Torque Teno Virus Load and Acute Rejection After Orthotopic Liver Transplantation. Transplantation 2017;101:e219–21.
- Mendez-Eirin E, Barge-Caballero E, Paniagua-Martin M, et al. Impact of Cytomegalovirus Infection on Long-Term Heart Transplant Outcome. J Heart Lung Transplant 2017; 36:S89.
- Sharon E, Shi H, Kharbanda S 4 et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLOS Comput Biol 2017;13:e1005629.
- Schütz E, Fischer A, Beck J, et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PLoS Med 2017;14:e1002286.
- Bloom RD, Bromberg JS, Poggio ED, et al. Cell-Free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221–32.
- Whitlam JB, Ling L, Skene A, et al. Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant. 2019;19:1037–49.
- Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor‐derived cell‐free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant 2019;19:3087–99.
- Gordon PM, Khan A, Sajid U,et al. An Algorithm Measuring Donor Cell-Free DNA in Plasma of Cellular and Solid Organ Transplant Recipients That Does Not Require Donor or Recipient Genotyping. Front Cardiovasc Med. 2016;3:33.
- Beck J, Bierau S, Balzer S, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem 2013;59:1732–41.
- Boer K, Baan CC, van Donk, N, et al. Donor-Derived Cell-Free DNA as Minimally Invasive Tool to Diagnose Acute Rejection after Kidney Transplantation. Transplantation 2018;102:S689.
- Zou J, Duffy B, Steward N, et al. Detection of donor cell-free DNA using digital PCR in lung transplant recipients with graft rejection and infection. Hum Immunol 2015;76:68.
- Tsuji J, Weng Z. Evaluation of preprocessing, mapping and postprocessing algorithms for analyzing whole genome bisulfite sequencing data. Brief Bioinform 2016;17:938–52.
- Lehmann-Werman R, Magenheim J, Moss J, et al. Monitoring liver damage using hepatocyte-specific methylation markers in cell-free circulating DNA. JCI insight 2018;21: pii: 120687.