Clostridioides (previously Clostridium) difficile has received much attention in the past couple of decades due to its rapid spread and rising virulence. C. difficile increasingly has been reported outside of acute care facilities in nursing homes and community home settings.
In 2013, the Centers for Disease Control and Prevention (CDC) listed C. difficile as one of three antimicrobial-resistant urgent threats—of the highest level of concern to human health (2). Also in 2013, the Centers for Medicare and Medicaid Services (CMS) mandated that healthcare institutions track and report C. difficile infection (CDI) rates to the National Healthcare Safety Network, which enables the public to compare rates between similar hospitals.
Debates about the most appropriate diagnostic method for CDI, as well as the investment needed to accurately capture CDI rates, have been heated within the laboratory and clinical community. This article will review factors affecting the choice of laboratory diagnostic methods for diagnosing C. difficile in light of the recent Infectious Diseases Society of America (IDSA)/Society for Healthcare Epidemiology of America (SHEA) C. difficile guidelines (1).
Clinical Disease States
C. difficile can cause life-threatening diarrhea and colitis (i.e., inflammation of the colon) primarily in individuals who recently have taken antibiotics and recently had contact with the medical system. An anaerobic Gram-positive bacillus, C. difficile possesses spores highly resistant to alcohol gels and many disinfectants that persist on inanimate surfaces for months if those surfaces are inadequately cleaned.
Once ingested, the spores can germinate in the intestine and produce toxins. The toxins can cause fluid accumulation in the bowel, leading to diarrhea or gastrointestinal complications. Infection control precautions must be instituted to curb the spread of the spores; thus, accurate and rapid diagnosis of C. difficile is critical.
C. difficile can exist within our bodies in various states. Consider the following scenario: A 92-year old nursing home resident was admitted with altered mental status, after having recently completed a 3-week course of levofloxacin for urinary tract infection. She had not had bowel movements for the past 2 days.
On admission, her peripheral white blood cell (WBC) count was 90,100 cells/mm3 (normal range 3,600–11,100 cells/mm3) with 24% bands (normal range 0–6%), or immature neutrophils. An abdominal computed tomography scan revealed that she had diffuse mucosal and bowel wall thickening, consistent with megacolon. A C. difficile test performed on rectal swab by toxin A/B enzyme immunoassay was positive. Subsequently this patient had a colectomy and started treatment with metronidazole.
Representative photos of a colectomy specimen obtained from a different patient with CDI demonstrate pseudomembranes visible on gross examination (Figure 2A) as well as microscopically (Figure 2B), which resembled this patient’s specimen.
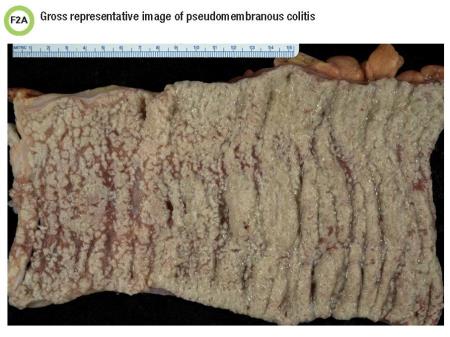
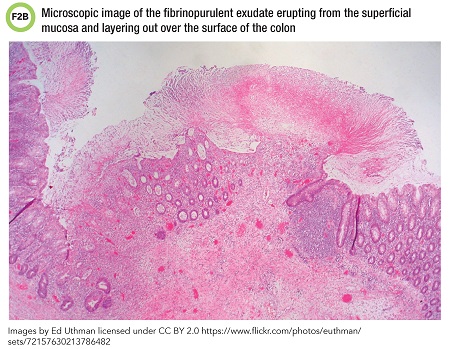
This scenario illustrates severe disease due to C. difficile. CDI may present with a range of clinical findings from diarrhea to pseudomembranous colitis to toxic megacolon. Sigmoidoscopy or histopathology may show thick, adherent layers of inflammatory cells and mucus, also known as pseudomembranes.
The most significant risk factor for CDI is antibiotic exposure. Although clindamycin, broad-spectrum cephalosporins, ampicillin, and fluoroquinolones typically are implicated in CDI, any antibiotic can cause this condition. When a person takes an antibiotic, beneficial bacteria in the intestine are destroyed or impaired for a period of time, thus increasing the likelihood that C. difficile may lead to infection.
Patients also can be colonized with or carry C. difficile without symptoms. This colonization state complicates clinical diagnosis, since the organism can be detected but isn’t necessarily causing disease. Colonizations with toxigenic (ie. toxin-producing) strains occurs in up to 15% of patients, and approximately 6% of patients are colonized with nontoxigenic strains. (3).
Consequently, merely detecting the C. difficile organism or its toxins within intestinal contents—stool or otherwise—does not mean they are causing disease. A patient’s clinical state at the time of sample collection determines whether CDI should be considered.
Pediatric CDI is more difficult to diagnose. Infant intestinal cells do not appear to have receptors for C. difficile toxins, so neonates may have detectable C. difficile in their stools without necessarily manifesting with disease. This makes the results of C. difficile testing difficult to interpret in children with diarrhea, and some laboratories do not perform testing for C. difficile for children younger than 1 or 2 years of age.
Laboratory Diagnostic Methods
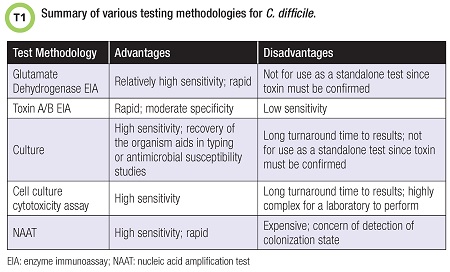
Several C. difficile diagnostic methods are available (Table 1). Test targets include the C. difficile organism itself or its toxins.
C. difficile produces a variety of toxins. Toxin A, encoded by the tcdA gene, is an enterotoxin that causes diarrhea. Toxin B, encoded by the tcdB gene, causes cellular distortion in vitro and presumably affects cells in vivo. The tcdC gene regulates toxin A and B production. Genes encoding for toxins A and B are present on the pathogenicity locus (PaLoc).
The NAP1/BI/027 strain was discovered in the early 2000s to produce a newly recognized toxin—the binary toxin (4). Although the function of the binary toxin has not been well elucidated, the cdtA and cdtB genes that encode it are located near the PaLoc. Assays target a variety of these genes and/or toxins.
Enzyme immunoassays (EIAs) detect toxins A and/or B, and some assays detect glutamate dehydrogenase (GDH), a conserved metabolic enzyme—a common antigen—secreted by C. difficile. Presence of GDH confirms presence of C. difficile but not necessarily its toxins. GDH EIAs detect the enzyme produced by both toxigenic and/or nontoxigenic strains of C. difficile. GDH is produced at much higher levels than toxins A and B.
A key advantage of GDH testing is its rapid turnaround time. Recent studies of this method also have demonstrated fairly high sensitivities (5). The disadvantage of GDH testing is that it can’t be used as a stand-alone test for CDI, and a confirmatory test for the presence of toxin is needed. These assays have been used in multistep and algorithmic approaches to diagnosis in combinations of testing involving toxin EIA, molecular assays, or other assays.
Toxin A/B EIAs target the toxins rather than C. difficile itself but are less sensitive than other methods in diagnosing CDI. However, these tests are rapid and may be combined with GDH targets to increase sensitivity.
Toxigenic culture is a highly sensitive method of recovering the organism when selective culture media are used. Recovering C. difficile strains allows for further molecular typing studies, such as to compare strains’ relatedness, or for antimicrobial susceptibility testing when indicated.
However the organism is recovered, presence of the toxin must be confirmed. Newer commercially available chromogenic media are also available to aid in organism recovery. Cell culture cytotoxicity assays may be performed to detect the toxin directly from stool. Time to results by this method is typically 24 to 48 hours.
Finally, molecular-based assays such as nucleic acid amplification tests (NAATs) detect the genes encoding for the toxins rather than the toxins themselves. Various NAATs are commercially available to laboratories, and these assays are highly sensitive.
Laboratory Diagnostic Testing Considerations
NAATs rose in popularity over the past 10 years based on their high sensitivity in detecting CDI. However, given concerns that NAATs may be detecting colonization states, some groups have advocated multistep or algorithmic approaches to diagnosis that make use of EIAs rather than relying solely upon molecular assays. No one answer provides the single, best approach for all institutions and hospitals; rather, laboratory approaches will vary based upon preanalytical issues of specimen type, the patient population under assessment, and the intended use of the test.
The type of specimen a laboratory receives is important when interpreting results of an assay. In most situations, only unformed stools should be tested when assessing for CDI. Formed stools may be tested, however, in cases of ileus or toxic megacolon when stool is not passed often, such as in the case we presented earlier, in which the patient was no longer producing stool due to infection-driven intestinal stasis. Given the colonization state associated with C. difficile organisms or its toxins, a positive C. difficile test in a person without diarrhea does not imply CDI.
Some laboratories encounter more stool collection-related issues than others. Diarrhea is defined as three or more unformed stools passed within the past 24 consecutive hours. This definition can be somewhat difficult to apply to certain patients who may not be able to relay this information to healthcare providers. Besides, the difference between unformed stools and formed stools may also be difficult to discern. Laboratory guidelines address what constitutes a formed stool—generally defined as a stool which takes the shape of its container (6).
However, many laboratories now utilize liquid-based stool collection devices, which make it difficult to ascertain the solidity of a stool. These devices are projected to become ever more popular as clinical microbiology laboratories embrace laboratory automation. Laboratories that utilize liquid collections must rely on clinicians to submit only loose stool specimens.
Electronic decision-making tools have been explored to decrease the probability that laboratories receive unacceptable specimens. Some institutions use an electronic order check that cross-references patients’ medical chart for use of laxatives or administration of other agents known to cause loose stools (7).
The recent 2017 IDSA/SHEA clinical practice guidelines for CDI suggest different approaches to laboratory testing based on specimens’ pre-test probabilities (1).
For instance, if clinicians institution-wide agree to screen carefully for clinical symptoms associated with CDI (i.e., at least three loose stools or unformed stools within 24 hours with a history of antibiotic exposure), then a highly sensitive test such as NAAT or a multistep algorithm may be best. Conversely, NAAT alone would not be the best choice for laboratories that operate without institution-wide criteria for stool submission and that typically receive unscreened stools for testing. These labs would do well to consider a multistep algorithm such as GDH plus toxin testing or NAAT plus toxin testing.
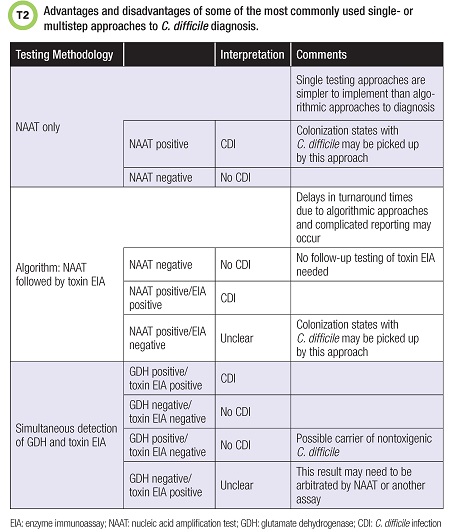
Laboratories may choose to utilize an algorithmic or multistep approach to CDI diagnosis with more than one testing methodology.
One common approach includes GDH testing combined with a toxin test, arbitrated by NAAT. Laboratories have many different approaches, but it is important to consider not only test performance but also other factors such as complicated algorithms for generating results when multistep testing methodologies are employed, as well as turnaround time (1). Table 2 lists some of the most commonly employed laboratory approaches to CDI diagnosis along with their advantages and disadvantages.
Labs also need to consider their patient populations when deciding which assays to use. Clinicians caring for patients with underlying gastrointestinal disorders that affect motility of the gastrointestinal tract have questioned the utility of relying only on a highly sensitive NAAT, since most of their patients suffer from loose stools at baseline.
The reasoning behind using a less sensitive assay for diagnosing CDI in this patient population presumably is that a toxin EIA would be less likely to detect C. difficile colonization than would a NAAT assay due to the lower sensitivity of the EIAs. The future of application of various C. difficile tests in different patient populations remains to be determined.
The final factor to consider when determining which assays to utilize for diagnosing C. difficile is the intent of testing.
Applying C. difficile testing as a screening test may require using a more sensitive detection platform than that used to diagnose C. difficile disease. Active screening for asymptomatic carriers has been explored recently owing to the belief that these patients may be reservoirs of bacteria, potentially transferring the organisms to other patients (8). Timely environmental cleaning and contact precautions are important steps if institutions use this approach.
Treatment and Control of CDI
The most significant risk factor for CDI is antibiotic exposure. Thus, providers begin therapy with an appropriate antimicrobial agent such as metronizadole or oral vancomycin and discontinue antimicrobial agents that may be predisposing to CDI. Best practices also call for contact precautions to be continued in patients with CDI for at least 48 hours after their diarrhea has resolved.
There is no clinical value in repeat CDI testing to establish whether a patient has been cured, as more than half remain positive for C. difficile even after successful treatment and resolution of diarrhea (1).
The Food and Drug Administration (FDA) approved in 2011 a macrolide antimicrobial, fidaxomicin, for the treatment of CDI, making it only the second agent after vancomycin approved for this purpose. The recent IDSA/SHEA guidelines for CDI treatment recommend using either vancomycin or fidaxomicin rather than metronidazole for an initial episode of CDI (1). If, however, the initial episode is not severe, metronidazole may be considered if access to vancomycin or fidaxomicin is limited.
More recently, FDA approved in 2017 bezlotoxumab, a human monoclonal antibody that binds to and neutralizes C. difficile toxin B, to prevent CDI recurrence in adults receiving antibiotic treatment. A single intravenous infusion of this compound was associated with a significantly lower rate of recurrent infection in clinical trials.
Conclusions
Accurate and rapid diagnosis of CDI is important for individual patients and for public health. C. difficile disease affects a large proportion of our population, with half a million infections per year and 15,000 deaths annually (2). The accuracy of testing methods depends on the methodologies used as well as pre-analytic factors related to specimen collection and the manner in which a test is used.
Effective laboratory testing requires close consultation among infection preventionists, clinicians, and laboratorians.
Audrey N. Schuetz, MD, MPH, D(ABMM), is an associate professor of laboratory medicine and pathology at Mayo Clinic College of Medicine and Science in Rochester, Minnesota. +Email: [email protected]
References
- McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018;66:987-94.
- Centers for Disease Control and Prevention. Antibiotic/antimicrobial resistance threats. https://www.cdc.gov/drugresistance/biggest_threats.html#cdiff (Accessed September 2018).
- Alasmari F, Seiler SM, Hink T, et al. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin Infect Dis 2014;59:216–22.
- Killgore G, Thompson A, Johnson S, et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol 2008;46:431–37.
- Eastwood K, Else P, Charlett A, et al. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol 2009;47:3211–7.
- Brecher SM, Novak-Weekley SM, Nagy E. Laboratory diagnosis of Clostridium difficile infections: There is light at the end of the colon. Clin Infect Dis 2013;57:1175–81.
- Truong CY, Gombar S, Wilson R, et al. Real-time electronic tracking of diarrheal episodes and laxative therapy enables verification of Clostridium difficile clinical testing criteria and reduction of Clostridium difficile infection rates. J Clin Microbiol 2017;55:1276–84.
- McDonald LC, Diekema DJ. Point-counterpoint: active surveillance for carriers of toxigenic Clostridium difficile should be performed to guide prevention efforts. J Clin Microbiol 2018;56:pii:e00782–18.