Blood group antigens are polymorphic residues of protein or carbohydrate on the red cell surface. They can provoke an antibody response in individuals who lack them, and some antibodies can lead to hemolytic transfusion reaction or hemolytic disease of the fetus/newborn (HDFN). Researchers have identified the molecular basis of many red cell blood group antigens, and an actively maintained database currently lists over 1,600 alleles of 44 genes (1). This mini-review describes the major applications of the explosion of knowledge in blood group genetics to the practice of blood banking and transfusion medicine (Table 1, 2–5).
Blood banks and clinical laboratories routinely type blood donors and patients for ABO and Rh(D), as these are the most critical antigens for safe transfusion. Laboratories generally do not type for minor antigens—of the Rh, Kell, Duffy, Kidd, and MNS systems—but they do screen plasma for antibodies against these antigens (antibody screen). If a patient’s antibody screen is negative, units for red blood cell (RBC) transfusion must be ABO- and Rh(D)-compatible. If an antibody to a clinically significant minor antigen is present, units must additionally lack the corresponding antigen.
The red cell phenotype is the complement of antigens on the red cell surface. In transfusion practice, this term refers to the status of clinically significant antigens other than ABO and Rh(D), typically some or all of the following minor antigens: C/c and E/e (Rh system); K/k (Kell system); Fya/Fyb (Duffy system); Jka/ Jkb (Kidd system); and M/N and S/s (MNS system).
Red cell phenotype testing of blood donors and donor RBC units is used to identify antigen-negative units for transfusion, usually for patients with red cell antibodies, but sometimes for transfusion-dependent patients without antibodies, to prevent alloimmunization. Phenotype testing is performed by serological typing with specific antisera using direct or indirect (antihuman globulin phase) hemagglutination.
Serological typing methods are simple, but they require reliable typing antisera, and typing for multiple antigens is labor-intensive. For example, to identify two compatible RBC units for a patient with anti-K, anti-Jka, and anti-S, it would be necessary to test approximately 20 random units.
Red cell phenotype testing on patients is used selectively, to supplement routine pre-transfusion testing. It is indicated for patients who have, or are at risk for making, multiple antibodies. Knowing which antigens are absent allows an assessment of the antibodies that the patient can make. The phenotype is then used to select matched, antigen-negative RBC units, thus avoiding exposure to foreign antigens and further alloimmunization.
However, standard serological typing cannot be used if the patient was transfused recently, because donor red blood cells can persist in the circulation for up to 3 months after transfusion. Also, standard serological typing cannot be used for many antigens if the patient has a positive direct antiglobulin test (DAT), as only antigens detectable by direct agglutination can be typed. Specialized serological methods can overcome these limitations but are not always successful.
Molecular Typing of Blood Donors
The expression of many clinically significant antigens is determined by single nucleotide polymorphisms (SNPs). Detecting these SNPs can predict the red cell phenotype and is an alternative to serological typing. Multiple SNPs can be included in a single assay, allowing efficient screening for multiple antigens. For this reason, molecular typing is eminently suitable for the mass screening of blood donors, and is expected to greatly expand the pool of blood donors (and donor RBC units) who are negative for multiple antigens or negative for a high-prevalence antigen.
Molecular typing also provides the means to identify antigen-negative donors when typing antisera are not available. For example, typing antisera for the Dob antigen (Dombrock group) are notoriously unreliable, and molecular typing for the DO gene mutation, 793A>G, that encodes the Dob antigen is a superior means of identifying Dob-negative donors for the patient with anti-Dob. This feature of molecular typing has also been applied to better characterize the reagent cell panels that are used for patient antibody identification.
Accurate typing of blood donors is essential to avoid immunizing potential recipients or precipitating a hemolytic transfusion reaction. A blood donor with weak antigen expression can immunize an antigen-negative transfusion recipient, and a significant advantage of molecular typing is its ability to identify antigenic variants. An example is the donor with Fyx, a weak form of Fyb, who may be mistyped as Fyb-negative by serology. Molecular typing can correctly identify this donor as Fyb-positive by demonstration of the 125A polymorphism that characterizes the FYB allele, and the 265C>T mutation that is responsible for weak antigenic expression.
Molecular typing can also help resolve typing discrepancies for other antigens, including ABO and D antigens. When multiple forms of a variant antigen are known to occur, molecular typing of donors and patients can potentially improve transfusion compatibility, because molecular typing can precisely identify the variant alleles even if they cannot be differentiated by serological typing. For example, a patient with anti-e associated with a variant e antigen should logically be transfused with donor units that are matched for her specific molecular variant. This approach of genotype-matching of donors and recipients is currently not practical, but it may become standard practice when molecular typing for clinical applications becomes broadly implemented.
Molecular Typing of Transfusion Recipients
Molecular typing is typically performed on DNA isolated from peripheral white blood cells. This sample is not affected by admixture with donor blood or by antibodies on the red cell surface, making molecular typing an excellent method to determine the phenotype when the patient is recently transfused or has positive DAT. The following cases illustrate these applications of molecular typing.
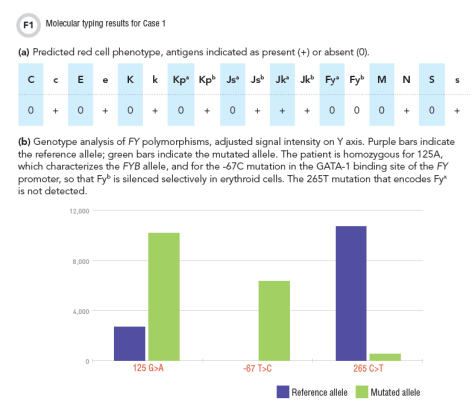
Case 1: An 18-year-old African-American male with sickle cell anemia, typed O positive and had anti-E antibody. As RBC transfusions must be compatible with the known antibody, the patient required E-negative RBC units. To reduce the risk of further alloimmunization, additional matching of the units to the patient’s red cell phenotype was desirable, but phenotype testing was precluded by transfusion 3 weeks earlier (6). Could molecular typing be used to predict this patient’s phenotype?
Molecular typing predicted the patient’s red cell phenotype as negative for C, E, K, Fya, Fyb, M, and S (Figure 1a). His Fyb phenotype was based on the presence of the FYB gene, in combination with the silencing mutation, -67T>C (Figure 1b). This mutation is commonly observed in individuals of African ancestry, and it silences Fyb expression only in erythroid cells: Fyb expression is unaffected in other tissues and the patient is not at risk for making anti-Fyb (7).
Based on the molecular typing results, antigen-negative RBC units were selected to be negative for C, E, and K, and when possible, also for Fya and S. Matching for M antigen was not indicated in this case because anti-M is usually not significant for transfusion.
Case 1 illustrates a common application of molecular typing on patient samples: red cell antigen typing when the patient’s phenotype cannot be tested by serological methods. However, this approach should be used cautiously, because the predicted phenotype may not always reflect the actual phenotype. Knowing the patient’s racial/ethnic background and the mutations included in the assay can be helpful in assessing the reliability of the results.
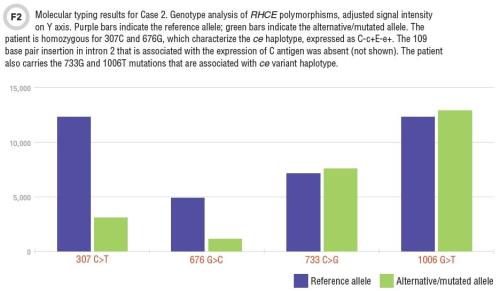
Case 2: A 42-year-old African-American man with renal failure, typed A positive and had anti-C antibody. His red cell phenotype was, unexpectedly, C+c+,E-, K-. Could molecular typing explain the inconsistency of anti-C antibody in this C-positive patient?
Variant Rh antigens are not uncommon among individuals of African ancestry, and should be considered if an antibody is noted against an antigen for which the patient is apparently positive. On molecular typing, this patient lacked the polymorphisms that encode the C antigen—he was truly C-negative. Interestingly, he carried the RHCE mutations, 733G and 1006T (Figure 2). These mutations suggest that the patient may carry a variant ce allele that is sometimes linked to a hybrid RHD allele; this hybrid RHD encodes a partial C antigen, and would explain the C-positive result on serological typing.
Indeed, further molecular testing demonstrated these variant alleles (RHCE*ceS allele, linked to RHD-CE(4-7)-D allele) (8). Thus, molecular testing was instrumental in determining the basis of the anti-C antibody in an apparently C-positive patient.
Case 2 illustrates a less common, but rapidly emerging application of molecular typing, in which identifying variant antigens provides information critical to patient management that would not be available from serological typing alone.
Molecular Typing in Prenatal Testing
Females of child-bearing potential with weak D, or discrepancies on D typing, are prime candidates for molecular typing, as this is the only means to distinguish between weak expression of D antigen (no risk of alloimmunization) and partial D antigen (with risk of anti-D alloimmunization).
Case 3: A 30-year-old Caucasian woman, G2P1, at 18 weeks of gestation, with no red cell antibodies, came to attention due to a typing discrepancy: she typed O negative at the current presentation, but was typed O positive during her last pregnancy 2 years ago. Standard prenatal practice dictates that Rh-negative prenatal patients be given Rh immunoglobulin to prevent anti-D alloimmunization, but in this case, the risk of anti-D alloimmunization is not clear. Could molecular typing determine the patient’s risk?
The RHD variants—weak D types 1, 2, and 3—are not associated with anti-D alloimmunization, and the aim of molecular typing in Case 3 was to determine if one of these variants was present. The patient was found to carry the 1154G>C mutation, which characterizes weak D type 1, a common variant among individuals of European ancestry. Based on this result, she was judged not to be at risk for anti-D alloimmunization and not a candidate for Rh immunoglobulin (9). On the other hand, if the weak D types 1, 2, or 3 mutations are absent in a prenatal patient with a D typing discrepancy, she may be assumed to be at risk for anti-D alloimmunization and will require prophylaxis with Rh immunoglobulin.
For the alloimmunized prenatal patient, molecular typing may be used to determine the antigen status of the fetus. For the common antigens except D, the father’s antigen type by serology is used to predict the antigen status of the fetus. If the father is antigen-negative (e.g., mother has anti-K, father types K-/k+), and paternity is certain, the fetus is expected to be antigen-negative also, and not at risk for HDFN from the mother’s antibody.
However, if the father’s antigen status is heterozygous (e.g., mother has anti-K, father types K+/k+), or unknown, the antigen status of the fetus cannot be inferred. It may be tested directly by molecular typing of fetal cells, usually amniocytes. Fetal antigen typing requires considerable knowledge and expertise, and must include evaluation for unusual variants, because failure to detect the gene in this setting has serious consequences. The results of testing must be evaluated in the context of parental results.
For the prenatal patient with anti-D, and a D-positive partner, serological testing cannot reliably determine zygosity of the paternal antigen, and molecular testing for RHD zygosity is indicated. This testing typically includes detection of the common mutations associated with absent D, such as RHD deletion, RHD pseudogene, and hybrid allele made up of portions of RHD and RHCE.
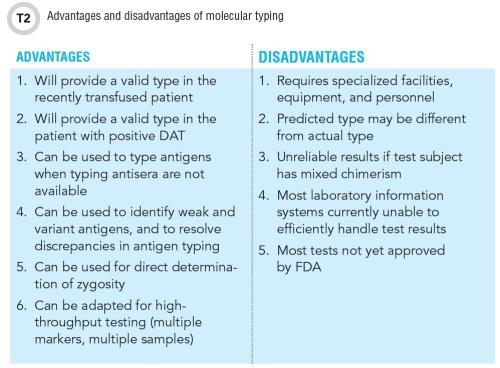
Disadvantages of Molecular Typing
Molecular typing determines the red cell phenotype when serological typing is unsuccessful or cannot provide the necessary information. But the disadvantages of molecular methods must be acknowledged in order to interpret and use the test results appropriately (Table 2).
Molecular typing results are affected by the quality of the DNA sample, and may be unreliable on samples with low DNA concentration. Results should be interpreted guardedly in stem cell transplant recipients, who may have more than one population of cells in peripheral blood. Typing on alternative samples such as a buccal smear may be helpful in interpreting such cases.
Molecular assays do not test for every possible polymorphism, and as previously noted, false prediction of positive antigen status can occur if a mutation that affects antigen expression is not represented on the assay, such as an unusual polymorphism or a mutation in a regulatory gene. This is of particular concern for individuals from a racial/ethnic minority group. If the predicted phenotype is inconsistent with the patient’s known antibodies or serological phenotype, the discrepancy must be investigated.
For example, if a patient with anti-Jka is predicted Jka-positive by molecular typing, the possibility of a variant Jka antigen should be considered. There are several examples of misleading results of molecular typing in the literature, and supplementation with serological typing is advised, whenever feasible.
Practical Aspects of Molecular Typing
Several SNP-based assays are commercially available, and one assay, based on bead microarray technology, was recently approved by the Food and Drug Administration for typing blood donors and transfusion recipients. Some assays may be suitable for implementation at a hospital laboratory, but for most institutions, it is more practical to access molecular typing through a reference laboratory. This approach offers the advantages of customized testing using multiple methods and platforms, and expert interpretation of results.
Molecular typing is costly, and results may not be available for days to weeks. It is not a substitute for serological testing and is best applied to patients for whom the results are expected to affect long-term management.
Conclusion
Serological testing remains the mainstay of the blood bank and transfusion medicine laboratory, but molecular typing is an invaluable supplement to traditional methods. Some administrative and logistical challenges to molecular typing remain unresolved, including high cost, slow turnaround, need for specialized equipment and technologists, and absence of information systems for handling the results. However, the development and marketing of molecular typing tests is progressing swiftly, and these tests are likely to become essential—rather than optional—for blood donor and patient testing. Widespread adoption of molecular testing will uncover novel solutions to transfusion problems, but improved detection of antigenic variants will surely pose new challenges for antigen matching (10).
References and Suggested Reading
1. Patnaik SK, Helmberg W, Blumenfeld OO. BGMUT: NCBI dbRBC database of allelic variations of genes encoding antigens of blood group systems. Nucleic Acids Res 2012 Jan;40:D1023–9
2. Denomme GA, Fernandes BJ. Fetal blood group genotyping. Transfusion 2007;47 Supplement:64S–8S.
3. Hillyer CD, Shaz BH, Winkler AM, et al. Integrating molecular technologies for red blood cell typing and compatibility testing into blood centers and transfusion services. Transfus Med Rev 2008;22:117.
4. Reid ME. Transfusion in the age of molecular diagnostics. Hematology Am Soc Hematol Educ Program 2009:171–7.
5. Denomme GA. Prospects for the provision of genotyped blood for transfusion. Br J Haematol 2013;163:3–9.
6. LaSalle-Williams M, Nuss R, Le T, et al. Extended red blood cell antigen matching for transfusions in sickle cell disease: A review of a 14-year experience from a single center. Transfusion 2011;51:1732–9.
7. Castilho L. The value of DNA analysis for antigens in the Duffy blood group system. Transfusion 2007;47:28S–31S.
8. Chou ST, Westhoff CM. The role of molecular immunohematology in sickle cell disease. Transfus Apher Sci 2011;44:73–9.
9. Flegel WA, Roseff SD, Tholpady A. Phasing-in RHD genotyping. Arch Pathol Lab Med 2014;138:585–8.
10. Chou ST, Jackson T, Vege S, et al. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood 2013;122:1062–71.
Suneeti Sapatnekar, MD, PhD, is a transfusion medicine staff physician at the Robert Tomsich Pathology and Laboratory Medicine Institute at the Cleveland Clinic in Cleveland, Ohio.
+Email: [email protected]