Scientists at Children's Mercy Hospital (CMH) in Kansas City recently reported being able to perform whole genome sequencing (WGS) and provide a provisional genetic diagnosis within 50 hours, far faster than the more typical 4- to 6-week or even longer sequencing and analysis cycle. This landmark proof-of-concept study gained world-wide attention and unquestionably moved the genomics field closer to making WGS practical for everyday clinical use. However, experts and even the research team itself caution that there are miles to go and many hurdles to clear before rapid WGS becomes a routine component of medical care. Even so, clinical laboratorians will do well to keep abreast of this fast-moving field so they can navigate the coming genomic world successfully.
"This research is helpful in a number of respects. It begins to show the outlines of how we can implement in healthcare this dramatic new technology, massively parallel sequencing, to make diagnoses and illuminate in at least a fraction of our patients what's going on clinically. The fact that they were able to do it quickly can be important especially in the context in which they were working," said James Evans, MD, PhD, Bryson distinguished professor of genetics and medicine at the University of North Carolina at Chapel Hill. "That said, we have to work within the confines of our ignorance and focus on what we know. The reality is that we can generate a whole exome or genome sequence, but we don't know how to interpret the vast majority of it, and we're not going to be able to fully interpret one anytime soon. The researchers did a nice job with their informatics feature of analyzing variants known to cause disease in newborns, the population they were working with. This is the kind of approach we'll need to use for the foreseeable future."
The 50-Hour Pathway
The study in question involved both retrospective and prospective 50-hour WGS (Sci Transl Med 2012;4:154ra135). In the retrospective work, the researchers confirmed previously diagnosed genetic diseases in two children. The prospective analysis involved five newborns with symptoms strongly suggestive of genetic disorders, and some of their family members. The researchers partnered with Illumina in Essex, England to use the company's HiSeq 2500 platform to sequence the genomes, which took 25.5 hours, plus 4.5 hours' sample preparation time. The 25.5 hours represented actual sequencing time, and did not include sample transport time from Kansas City to England.
The project also involved two CMH-developed software tools, symptom- and sign-assisted genome analysis (SSAGA), and rapid understanding of nucleotide variant effect software (RUNES), to automate and speed up the variant calling and analysis process. SSAGA and RUNES also helped focus the analysis, as recommended by guidelines for genetic testing in children, on areas of the genome associated with variants relevant to the babies' symptoms. With these software tools, the process of base and variant calling, sequence alignment, and variance interpretation took 20 hours. (See Box, below, for summary of the WGS timeline.)
Counting Down to the 50-Hour Genome
Researchers at Children's Mercy Hospital (CMH) in Kansas City recently reported performing rapid whole genome sequencing on newborns in CMH's neonatal intensive care unit who had symptoms strongly suggestive of genetic disorders. Using both the latest sequencing technology and sophisticated software tools, the research team was able to sequence the babies' DNA and arrive at a provisional genetic diagnosis within 50 hours. Here is the breakdown in time to achieving this milestone.
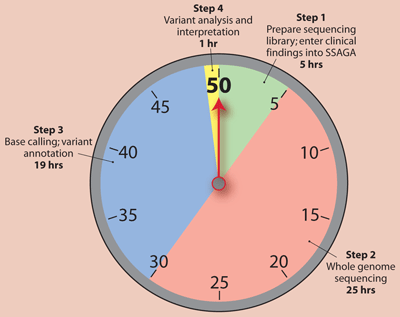 Legend: SSAGA, symptom- and sign-assisted genome analysis
Legend: SSAGA, symptom- and sign-assisted genome analysis
Adapted from Sci Transl Med 2012;4:154ra135
The rapid WGS yielded approximately 40-fold aligned genomic coverage, with >99.5% concordance between variant genotypes and those detected via deep sequencing. The prospective WGS led to a definitive or likely molecular diagnosis in four of five babies. The researchers confirmed each diagnosis via Sanger sequencing in CMH's CLIA lab, which took 4 days.
Emergency Genomics
The research team, led by Stephen Kingsmore, MB, ChB, DSc, FRCPath, views its effort as an early prototype for the use of rapid WGS in neonatal intensive care units (NICU)—the epicenter of genetic disease presentations. More than 20% of infant deaths result from genetic diseases, and a sizeable number of the 3,528 Mendelian diseases already are manifest, to greater or lesser degrees, at birth. Newborn screening, though rapid and cost-effective, identifies a mere pittance of these disorders. This leaves clinicians frustrated and parents anguished about what exactly is wrong with acutely ill babies, many of whom are discharged or die without a definitive diagnosis.
In this context, rapid diagnosis could enable clinicians to implement effective treatments early on as well as to discontinue further futile diagnostics and treatments. Families could be spared expensive and emotionally draining diagnostic odysseys to uncover the cause of their children's problems. Further, even if rapid WGS were to remain a relatively expensive test with sequencing and interpretation costing, by CMH's estimate, $13,500, it still would be a bargain in comparison to multiple and/or lengthy hospitalizations, perhaps opening the door for payer coverage of this new technology. For all these reasons, Kingsmore argued that the NICU perhaps is uniquely suited for rapid WGS and was circumspect about how much a harbinger the team's achievement might be for other clinical settings.
"The novel idea that we introduced was the concept of emergency genomics in which even in acute clinical situations we could start to give answers in a time frame that can guide medical decisions. About three to four percent of children have a genetic disease and 15 percent of hospitalized kids have a single-gene disease. We believe that as many as 30 percent of NICU admissions would benefit from genomic analysis, so we're very focused on this huge area of diagnostic need," he said.
Kingsmore went on to add that with decades of genetic testing under its belt, the field finally is developing technologies to better and more efficiently tackle analysis of the 3.4 million variants in each genome, to the point of making that information relevant to acute clinical management. "In our case, just making a prototypic system was a huge hurdle. Getting a body of evidence that genomes truly impact NICU care and starting to understand where this is cost-effective, those are still really difficult tasks ahead of us that will likely take a couple of years. There are several other areas of medicine where emergency genomes are likely to be incredibly powerful, and those applications are on the radar screens of other groups, not ours. This is a big urgent problem and we all still have a heck of a lot of work to do." Kingsmore is director of CMH's Center for Pediatric Genomic Medicine.
Among other things, CMH is still shaving down the time. When CLN spoke with Kingsmore, he said it might soon be possible to shorten the process to 24 hours. "Getting it to one day is more difficult, but I'd love to be able to do it by the end of 2013," he said. "I'm really looking forward to the day when we can decode a patient's genome in a couple of hours at a cost that's not prohibitive. That's the magical thing. You visit the doctor, and later that day he or she can call you back with the results. That's going to take a long time and a lot of thought as to how to communicate genome information, but that's the line we're on."
For Neonates Only?
Though upbeat about the CMH team's general approach, Evans sounded a cautionary note about other near-term clinical applications for rapid WGS. "We have to resist the nonsensical but somewhat seductive idea that everybody is going to benefit from whole genome sequencing and that everybody should have it," he observed. "Kingsmore's group picked a good patient population enriched for genetic conditions in which these results matter, because in some cases it changed treatment decisions. However, most patients with a genetic condition are not in a circumstance that is as time sensitive. As you get on in life, fewer diseases are caused primarily by genetics."
Other experts viewed the study as a realistic peak into how rapid WGS might be implemented outside NICU. "This is a model for pushing the limits of how fast testing can get done. Of course in the NICU setting it's much more critical to get results back fast, but it won't be far along that people will be thinking about this for prenatal testing," predicted Chrisa Lese Martin, PhD, FACMG, associate professor of genetics at Emory University School of Medicine in Atlanta. "Even in the pediatric setting there could be a role for this quick turnaround time, for example in children who present with intellectual disabilities or autism. The time to results in many labs is four-to-six months. That's a long time to wait, both for the families and for clinicians, without being able to act if something's there. A lot of these could be inherited mutations and a family could get pregnant again before they get results back."
Two influential groups, the American Academy of Pediatrics and the American College of Medical Genetics, recently updated long-standing policies about genetic testing in children. Both oppose carrier screening in children when this testing does not provide health benefits in childhood (See Box, below).

Spurring Others On
Whether or not the CMH team's achievement has immediate broader clinical applications, it has inspired other research teams to press the testing and analysis envelope. "The Kingsmore team did the experiment that captured the field's imagination. It was something many of us thought could be done, but they executed, and for that they should be commended," said David Craig, PhD, deputy director of bioinformatics at Phoenix-based Translational Genomics Research Institute (TGen).
That sentiment was echoed by Heidi Rehm, PhD, FACMG, clinical laboratory director for the Partners Healthcare Center for Personalized Genetic Medicine in Boston. "One of the challenges for those of us trying to offer useful clinical genomic sequencing is turnaround time. Right now, it often takes 60 to 90 days to return the results of a whole genome sequence, and if having that sequence is going to make a difference in how you manage a child with a suspected genetic disorder, then waiting that long is not consistent with the notion of this information being useful in a clinical care context," she said. "This proof-of-concept study was important in showing us it can be done rapidly, and in saying it's not just something we can do at our leisure, but in real time in the course of clinical management of patients."
Exposing the Weak Spot
Though he was positive about CMH's work, Craig also contended the team's report underscored the genomic field's weak spot: informatics. That is not to say SSAGA and RUNES are not innovative. Far from it. SSAGA enables any physician, not necessarily a clinical geneticist, to check off a sick baby's symptoms from a menu of 227 clinical terms arranged by nine symptom categories. SSAGA then nominates diseases and related genes associated with those symptoms, enabling the CMH team to avoid the thorny ethical issue of incidental findings in children that may have no relevancy in their lives for decades to come. This process also narrows substantially the variants that need to be analyzed. At the time CMH's study was published, SSAGA had been mapped to 591 genetic diseases, but the team has been steadily adding to it, with the goal of incorporating all 3,568 Mendelian diseases. The team also validated SSAGA by retrospectively entering the presenting symptoms of 533 children previously diagnosed with genetic disorders. It proved to have 99.3% sensitivity in proposing correct diseases and genes.
RUNES automates several steps in variant annotation to estimate the functional consequences of variants, enabling the interpretation to focus on variants known or likely to be pathogenic. It was validated with extensive automated testing.
These sophisticated tools were developed over a period of about 18 months in a collaborative effort led by CMH's director of informatics, Neil Miller. They are specific to CMH, which has no immediate plans either to commercialize or open the software to other users, contended Kingsmore. Therein lies the issue, according to Craig. "Every group working in this area has a software that is being innovative on pretty much the exact same thing. This is the problem with the field because these softwares are all home brew, and they're only being used in the place where they were developed," he explained.
Solutions on the Horizon
If Craig sees drawbacks with the field's fragmented software, he also sees solutions coming rapidly. One is the burgeoning number of specialty companies, many backed by venture capital, that are offering both á la carte and soup-to-nuts WGS and interpretation. One example out of many is SVBio, which in January announced a strategic collaboration with Mayo Clinic in which SVBio will provide clinical genomic interpretation services and clinical decision support interfaces to Mayo.
Another model for breaking out of the build-your-own genomic analysis software is Partner's GeneInsight and its newly formed GeneInsight Network, a collaboration with Illumina in which a group of leading commercial and academic labs plan to share knowledge about genetic variants and the cases in which they are observed. GeneInsight already offers to other labs its genetic analysis, interpretation, and reporting software, which is registered with the U.S. Food and Drug Administration as a Class I exempt medical device. "We believe the best strategy is in essence a crowd-sourcing strategy in which labs who are each curating knowledge agree to share it in a share-and-share-alike model massively increasing each lab's access to interpreted variants," said Rehm.
Reimbursement: The Inflection Point
These models parallel case-by-case inroads being made with payers to reimburse WGS testing that has immediate patient management consequences, something Craig said is happening more and more. "We're starting to see reimbursement occur with preauthorization and with some insurers. When genomic testing gets attached to reimbursement, that will be the inflection point where things start to change. That will move genomic testing past funding from the National Institutes of Health, philanthropy, and venture capital dollars to become part of healthcare," he predicted.
In this brave new world, experts see a crowded field of players and also heavy ongoing investments in technology and staff for any organization that seeks a go-it-alone strategy, all before WGS is even widely adopted in clinical care or recognized by payers. This holds even for groundbreaking CMH, which purchased a HiSeq 2500 this fall with philanthropic support and is continuing to perform WGS in selected neonates, but only as a research service also supported by private funding.
Where does this leave the average clinical lab? Martin advised laboratories not directly involved in genomic analysis to know about the labs they're sending genomic tests to. "They need to know what part of the work's being done by which lab, all of it, technical and interpretive, or only interpretive. Some labs only do the technical component and have another do the interpretation or vice versa. But it's part of the responsibility of the individuals ordering the test to ensure this testing is being done by appropriate centers adhering to College of American Pathologists and CLIA regulations," she said.
Miller and Evans emphasized the need for laboratories to collaborate closely with clinicians around both the translation and clinical adoption of genomic advances. Rehm urged laboratory professionals not to sit around waiting for the industry to mature. "The genomic space is moving quickly and soon we'll be in a world where it's a routine component of healthcare. Hospitals, labs, and clinics all need to have a plan in place for how they will interface with the world of genomic medicine, which is coming quickly."