
According to the American Cancer Society, prostate cancer (PCa) is the second most common type of cancer found in American men and the second leading cause of cancer death after lung cancer, with approximately 241,000 new cases reported in 2011. It is estimated that annually one in six men will be diagnosed with PCa, yet only 1 in 36, or approximately 34,000, is expected to die of the disease. Because PCa is typically not lethal, more men die with their disease than from it, making PCa management costly.
Today, blood tests for prostate specific antigen (PSA) are widely used to screen for PCa. This article will describe the current controversies in using this tumor biomarker and introduce two very different novel markers for detecting PCa: a proPSA isoform of free PSA, and PCA3, a nucleic acid marker.
The PCa Diagnostic Dilemma
While detecting PCa benefits many men, the blood test most frequently used to detect the disease, PSA, is less than perfect. The controversy over PSA screening was rekindled recently when The U.S. Preventive Services Task Force released an updated statement on PSA testing for men with a "D" rating, recommending for the first time that healthy men avoid getting regular PSA tests (1).
The reasons for this controversy are many; however, much attention has been focused on the limited specificity of PSA for detecting PCa, especially significant PCa. The latter is defined as a cancer that will shorten a man's life and typically has a Gleason Score (GS) of 7 or greater. Depending on the clinical setting, the cancer positivity rate of typical prostate biopsies from men with elevated PSA levels is only 20–40%. In other words, as many as four out of five prostate biopsies result in negative findings for PCa.
In fact, every year in the U.S. as many as 750,000 prostate biopsies are negative. These negative biopsies come with significant cost to the healthcare system, as well as considerable medical risks to the patient. For example, rectal bleeding following biopsy is fairly common, and the procedure itself poses a significant risk for infection. Furthermore, patients increasingly are infected with fluoroquinolone-resistant E. coli (2).
When a prostate biopsy is positive for cancer, the dilemma becomes how to appropriately manage the man's treatment. Certainly, a high-grade cancer (GS ≥7) should prompt consideration of aggressive treatment options. But the decision whether to treat a low-grade cancer, such as a GS 6, is less clear. Even some GS 7 cancers can be appropriately managed with conservative treatment. Many of these cancers do not kill men over a 10–20 year time period, and physicians have many treatment options available. Therefore, the challenge is how to make PSA a better marker for significant PCa.
PSA: The First Biomarker for PCa
In 1986, the Food and Drug Administration (FDA) first approved PSA as a biomarker for monitoring men with PCa. Shortly thereafter, it gained popularity as an aid in detecting PCa and was approved in 1994 for PCa screening. As the limitations of PSA as a PCa detection tool became increasingly apparent, however, researchers and clinicians explored derivatives of PSA in an attempt to increase the marker's specificity.
Further research showed that the rate of PSA increase—the velocity or doubling time—appeared to be an important indicator of significant disease. Researchers also looked at the percent of free PSA as a function of total PSA as a means to improve cancer specificity. Even after considerable research, the diagnostic performance of PSA today does not match, for example, that of cardiac troponin in diagnosing acute myocardial infarctions.
Does PSA Screening Save Lives?
Given this controversy, it's important to ask if PSA screening save lives. In general, the assumption is that screening saves men's lives by prompting earlier therapy. A number of modestly powered studies have in fact demonstrated that PSA increases the number of cancers detected while they are still localized, thereby increasing life expectancy for men with the disease.
In the 2000s, however, investigators initiated several larger efforts to determine whether a systematic PSA screening program decreases PCa mortality in men. Two such studies were simultaneously reported in April 2009 in the New England Journal of Medicine.
The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial was a large population based randomized trial designed and sponsored by the National Cancer Institute (NCI) to determine the effects of screening on cancer-related mortality and secondary endpoints in men and women age 55 to 74. This multi-centric study failed to demonstrate a mortality benefit for PSA screening (3).
In fact, the authors reported that the rate of deaths did not differ between the screened and non-screened groups. Unfortunately, the control (non-screened) arm of the study contained a large number of men who had received prior PSA testing, which created a contamination bias underestimating the benefit of screening. Furthermore, some subjects in the screened group had actually received PSA tests prior to the study, potentially skewing the results because prostate cancers could have been detected previously.
The second multi-centric study, the European Randomized Study of Screening for Prostate Cancer (ERSPC), began in the 1990s and involved more than 160,000 men from seven countries (4), with a mean follow-up of 9 years. This study also suffered from some flaws in the control arm, although far less so than in the U.S. trial. The investigators concluded that PSA-based screening reduced the rate of death from PCa by 20% but was associated with a high risk of over-diagnosis.
It now appears that the first report from the ERSPC may have underestimated the benefit of PSA screening. In a recent update, researchers found that a longer follow-up time and a younger screening population improved mortality estimates (5). In fact, in this tightly controlled, prospective study, PCa mortality was reduced by 29% over 11 years after adjustment for noncompliance. However, the study found no significant difference in overall or all-cause mortality.
An essential piece missing in all of these investigations is whether PSA screening reduces the morbidity of PCa, leaving unanswered the question of whether men live healthier lives because their PCa was detected and appropriately treated early. Alternatively, it also is important to ask if a positive PSA test results in overtreatment of PCa, allowing men to live longer, but lower quality, lives. These will be vital considerations in future research.
The Hunt for a Better Screening Biomarker
Despite decades of research, no clear consensus has emerged on the benefits of PSA screening. Further prospective studies are needed to settle the important issues outlined here. In light of the limitations of PSA as a PCa detection tool, much effort has been focused over the years on alternate biomarkers to supplement, or even replace the PSA test, as a tumor detection tool.
Even with the enormous research efforts focused on this disease, investigators have not uncovered an abundance of viable biomarkers for early detection of PCa. Recently, however, several novel markers have gained increased attention for their ability to supplement information from initial PSA test results. While not yet able to completely replace PSA as a screening tool, at least two markers appear to provide significant information for clinicians making patient management decisions for men with a modestly elevated PSA of 4–10 ng/mL.
Isoforms of Free PSA
Soon after PSA was reported to be a potential marker for PCa, researchers discovered that PSA exists in two different forms in serum: a complex of PSA and alpha-1-antichymotrypsin (complexed PSA), and a free form, not bound to an inhibitor (free PSA) (6). Researchers demonstrated that free PSA, expressed as a percentage of total PSA, was higher in benign tumors and lower in cancerous tumors, thereby improving the biomarker's specificity for PCa.
Further research showed that free PSA is not one, but at least three different molecules, all enzymatically inactive (Figure 1). One form, BPSA, is degraded at two specific sites on the protein (amino acids 145 and 182) and is more closely associated with benign prostatic hyperplasia (7). A second, poorly characterized form is an intact protein, but it is denatured and therefore enzymatically inactive. This form is known as intact PSA (non-native, non-nicked) (8). A third form is the pro-enzyme form of PSA, termed proPSA. Native proPSA contains a seven amino acid leader sequence that HK-2 and other activating enzymes cleave off to produce active PSA. Degraded forms of proPSA also exist with pro-sequences of five-, four-, and two-amino acids (7). While its function is not fully understood, [-2]proPSA is more highly associated with the peripheral zone of the prostate gland in cancer tissue. Researchers also have demonstrated that the protein can be accurately and reliably measured.
Figure 1
Typical Proportions of PSA and Free PSA Isoforms in
Prostate Cancer Serum
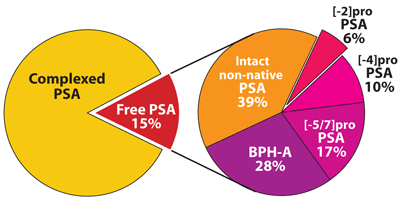 Redrawn from Kiyo J. J. Med 2003;52:86–91.
Redrawn from Kiyo J. J. Med 2003;52:86–91.
[-2]proPSA also looks promising as a tool for screening men for PCa. Similar to free PSA, which has no discriminating value by itself, [-2]proPSA demonstrates better performance when combined with total PSA and free PSA in a multi-marker index. In June, FDA approved Beckman Coulter's Prostate Health Index, or phi, which is indicated for use in men with a PSA of 4–10 ng/mL.
A multi-center study compared the ability of the phi index to discriminate PCa from benign disease in comparison to total PSA or %free PSA (9). The researchers evaluated prostate biopsy results from 892 men and compared them to results of PSA, free PSA, [-2]proPSA, and their derivatives, including phi. Using receiver operating characteristic (ROC) curve analysis, the area-under-the-curve (AUC) was 0.703 for phi index, 0.648 for %free PSA, and 0.525 for total PSA. At a fixed detection rate of 95%, phi increased the specificity for PCa 1.9-fold over % free PSA and 2.9-fold over total PSA. The phi index also demonstrated an association with the GS on prostate biopsy, with the probability of GS ≥7 increasing as the proPSA index increased.
PCA3 RNA
Proteins are not the only type of marker being investigated for more specific detection of PCa. Researchers identified prostate cancer gene 3 (PCA3), first known as DD3, using differential display analysis to compare the mRNA expression patterns of normal versus tumor tissue of the human prostate. PCA3 is a prostate-specific, non-coding mRNA that is overexpressed in 95% of prostate cancers tested, with a median 66-fold up-regulation compared with adjacent non-neoplastic prostatic tissue (10).
The assay for the PCA3 gene is based on in vitro nucleic acid amplification of the mRNA. It uses a urine sample collected after an "attentive" digital rectal exam that has been mixed with an equal volume of urine transport medium to lyse the prostate cells and stabilize the mRNA. Following isolation from the prostatic epithelial cells, the PCA3 mRNA is amplified and quantified. PSA mRNA in the sample serves as both a control of adequate sampling and as a normalizing factor, since the PCA3 score is calculated as the ratio of PCA3 mRNA molecules to PSA mRNA molecules. Unlike serum PSA, the PCA3 score is not correlated with prostate volume.
Using a prototype assay for PCA3, researchers performed a ROC curve analysis of pre-biopsy specimens of a population with PSA results of 7.7 ng/mL (mean) and 35.9 ng/mL (97.5 percentile). PCA3 produced an AUC of 0.746, with sensitivity of 69% and specificity of 79%. In this study, PCA3 and PSA mRNAs were not detectable in specimens from patients following prostatectomy, except for one man with recurrent prostate cancer.
A number of investigators have evaluated the PCA3 urine test in men undergoing repeat or initial prostate biopsies (11–15). These studies demonstrated that PCA3, used in conjunction with serum PSA and other clinical information, can enable more accurate prediction of prostate biopsy outcome.
In the largest published study to date, PCA3 scores were measured prior to year 2 and year 4 prostate biopsies from a subset of 1,140 subjects in the placebo arm of the Reduction by Dutasteride of Prostate Cancer Events (REDUCE) trial, a prostate cancer risk reduction study of men with modestly increased PSA results (0.30 to 33.9 ng/mL) and negative prostate biopsy findings at baseline (11). The researchers found increasing levels of PCA3 mRNA were associated with increasing positive prostate biopsies findings, and that PCA3 results at year 2 were a significant predictor of year 4 biopsy outcomes (AUC 0.634, p <0.0002), whereas serum PSA and %free PSA were not predictive.
Earlier this year, FDA approved the first molecular test based on PCA3, the PROGENSA PCA3 test, intended to aid in the decision for repeat biopsy in men 50 years of age or older who have had one or more previous negative prostate biopsies. Developed by Gen-Probe, the test represents the latest PCa marker aimed at fortifying the use of PSA and its derivatives. The pivotal study for PROGENSA PCA3 involved 495 men at 14 clinical sites and only included men who were recommended for repeat biopsy. The PROGENSA PCA3 demonstrated a negative predictive value of 90%, meaning that a negative PROGENSA PCA3 assay result predicted a prostate biopsy negative for PCa 90% of the time.
The Search Goes On
Today, after many years of experience and extensive research, PCa remains a challenging disease to detect. We have long recognized that PSA as a screening biomarker has limitations, and that over-diagnosis and over-treatment of PCa are common. At least for now, it appears that PSA testing is the most effective biomarker available for helping to detect PCa.
Although PSA detects low-grade or indolent cancers, aggressive cancers pose a significant risk to men. Because of these challenges in the contemporary screening environment for PCa, the detection power of PSA might be better by combining information with emerging markers for PCa as described here. The experience with free PSA taught us that multi-marker strategies may have value in PCa detection. The [-2]proPSA assay, as part of the multi-marker phi index, is the next example of combining markers for increased utility. Also potentially important, but not covered in this article, are the markers known as PTEN, ANXA3, as well as gene fusion products such as TMPRSS2-erg.
Future research will undoubtedly focus on combinations of protein and nucleic acids markers to add to the initial information provided by total PSA. It will take time for physicians to gain comfort in making decisions with these new tools, but clearly there is a real need for better screening biomarkers for PCa.
REFERENCES
- Moyer VA. Screening for Prostate Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med 2012;157:1–15.
- Liss MA, Chang A, Santos R. Prevalence and significance of fluoroquinolone resistant Escherichia coli in patients undergoing transrectal ultrasound guided prostate needle biopsy. J Urol 2011;185:1283–1288.
- Andriole GL, Crawford, ED, Grubb, RL. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med 2012;360:1310–1319
- Schroeder FH. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 2009;360:1320–1328.
- Schroeder FH. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981–990.
- Stenman U-H. Serum concentrations of prostate specific antigen and its complex with alpha-1-antichymotrypsin before diagnosis of prostate cancer. Lancet 1994; 344:1594–1598
- Mikolajczyk SD. Free prostate-specific antigen in serum is becoming more complex. Urology 2002;59:797-802.
- Lilja H. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring Nature Reviews Cancer 2008;8:269–278.
- Catalona WJ. A multicenter ctudy of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/mL prostate specific antigen range. J Urol 2011;185:1650–1655.
- Groskopf J. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin Chem 2006;52:1089–1095.
- Aubin SMJ. Prostate cancer gene 3 score predicts prostate biopsy outcome in men receiving dutasteride for prevention of prostate cancer: Results from the REDUCE Trial APTIMA PCA3 molecular urine test. Urology 2011;78:380–385.
- Haese A, de la Taille A, van Poppel H, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. European urology 2008;54:1081–1088.
- Marks LS, Fradet Y, Deras IL, et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology 2007;69:532–535.
- de la Taille A, Irani, J, Graefen M, et al. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J. Urology 2011;185:2119–2125.
- Deras IL, Aubin SMJ, Blasé A, et al. PCA3: A molecular urine assay for predicting prostate biopsy outcome. J Urology 2008;179:1587–1592.

Bernard C. Cook, PhD, DABCC, FACB is Manager, Global Scientific Affairs, Beckman Coulter, Chaska, Minn.
Email